Abstract
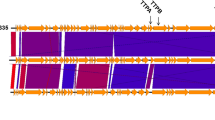
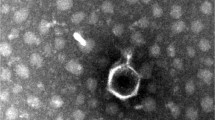
Carbapenem-resistant Klebsiella pneumoniae (CRKP) has spread globally and emerged as an urgent public health threat. Bacteriophages are considered an effective weapon against multidrug-resistant pathogens. In this study, we report a novel lytic phage, kpssk3, which is able to lyse CRKP and degrade exopolysaccharide (EPS). The morphological characteristics of kpssk3 observed by transmission electron microscopy, including a polyhedral head and a short tail, indicate that it belongs to the family Podoviridae. A one-step growth curve revealed that kpssk3 has a latent period of 10 min and a burst size of 200 plaque-forming units (pfu) per cell. kpssk3 was able to lyse 25 out of 27 (92.59%) clinically isolated CRKP strains, and it also exhibited high stability to changes in temperature and pH. kpssk3 has a linear dsDNA genome of 40,539 bp with 52.80% G+C content and 42 putative open reading frames (ORFs). No antibiotic resistance genes, virulence factors, or integrases were identified in the genome. Based on bioinformatic analysis, the tail fiber protein of phage kpssk3 was speculated to possess depolymerase activity towards EPS. By comparative genomics and phylogenetic analysis, it was determined that kpssk3 is a new T7-like virus and belongs to the subfamily Autographivirinae. The characterization and genomic analysis of kpssk3 will promote our understanding of phage biology and diversity and provide a potential strategy for controlling CRKP infection.


Similar content being viewed by others
Data availability statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Chen L, Kreiswirth BN (2018) Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis 18:2–3
Paczosa MK, Mecsas J (2016) Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661
Bengoechea JA, Sa Pessoa J (2019) Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev 43:123–144
Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY (2012) Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis 12:881–887
Huang W, Qiao F, Zhang Y, Huang J, Deng Y, Li J, Zong Z (2018) In-hospital medical costs of infections caused by carbapenem-resistant Klebsiella pneumoniae. Clin Infect Dis 67:S225–S230
Lin YT, Su CF, Chuang C, Lin JC, Lu PL, Huang CT, Wang JT, Chuang YC, Siu LK, Fung CP (2019) Appropriate treatment for bloodstream infections due to carbapenem-resistant klebsiella pneumoniae and Escherichia coli: a nationwide multicenter study in Taiwan. Open Forum Infect Dis 6:ofy336
Kelesidis T, Karageorgopoulos DE, Kelesidis I, Falagas ME (2008) Tigecycline for the treatment of multidrug-resistant Enterobacteriaceae: a systematic review of the evidence from microbiological and clinical studies. J Antimicrob Chemother 62:895–904
Wang X, Wang Q, Cao B, Sun S, Zhang Y, Gu B, Li B, Liao K, Zhao F, Jin L, Jin C, Yang C, Pei F, Zhang Z, Wang H (2019) Retrospective observational study from a chinese network of the impact of combination therapy versus monotherapy on mortality from carbapenem-resistant enterobacteriaceae bacteremia. Antimicrob Agents Chemother. https://doi.org/10.1128/AAC.01511-18
Zhang R, Dong N, Huang Y, Zhou H, Xie M, Chan EW, Hu Y, Cai J, Chen S (2018) Evolution of tigecycline- and colistin-resistant CRKP (carbapenem-resistant Klebsiella pneumoniae) in vivo and its persistence in the GI tract. Emerg Microbes Infect 7:127
Gordillo Altamirano FL, Barr JJ (2019) Phage therapy in the postantibiotic era. Clin Microbiol Rev. https://doi.org/10.1128/CMR.00066-18
Tang C, Deng C, Zhang Y, Xiao C, Wang J, Rao X, Hu F, Lu S (2018) Characterization and genomic analyses of pseudomonas aeruginosa podovirus TC6: establishment of genus pa11virus. Front Microbiol 9:2561
Monteiro R, Pires DP, Costa AR, Azeredo J (2019) Phage therapy: going temperate? Trends Microbiol 27:368–378
Górski A, Międzybrodzki R, Węgrzyn G, Jończyk-Matysiak E, Borysowski J, Weber-Dabrowska B (2019) Phage therapy: current status and perspectives. Med Res Rev. https://doi.org/10.1002/med.21593
Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S (2005) Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol 43:4178–4182
Yang C, Wang H, Ma H, Bao R, Liu H, Yang L, Liang B, Jia L, Xie J, Xiang Y, Dong N, Qiu S, Song H (2018) Characterization and genomic analysis of SFPH2, a novel T7virus infecting shigella. Front Microbiol 9:3027
Guo Z, Huang J, Yan G, Lei L, Wang S, Yu L, Zhou L, Gao A, Feng X, Han W, Gu J, Yang J (2017) Identification and characterization of Dpo42, a novel depolymerase derived from the Escherichia coli phage vB_EcoM_ECOO78. Front Microbiol 8:1460
Tang F, Zhang P, Zhang Q, Xue F, Ren J, Sun J, Qu Z, Zhuge X, Li D, Wang J, Jiang M, Dai J (2019) Isolation and characterization of a broad-spectrum phage of multiple drug resistant Salmonella and its therapeutic utility in mice. Microb Pathog 126:193–198
Yang Z, Liu X, Shi Y, Yin S, Shen W, Chen J, Chen Y, Chen Y, You B, Gong Y, Luo X, Zhang C, Yuan Z, Peng Y (2019) Characterization and genome annotation of a newly detected bacteriophage infecting multidrug-resistant Acinetobacter baumannii. Arch Virol 164(6):1527–1533
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Borodovsky M, Lomsadze A (2014) Gene identification in prokaryotic genomes, phages, metagenomes, and EST sequences with GeneMarkS suite. Curr Protoc Microbiol. https://doi.org/10.1002/0471250953.bi0405s35
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD (2012) The Pfam protein families database. Nucleic Acids Res 40:D290–D301
Liu B, Zheng D, Jin Q, Chen L, Yang J (2019) VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692
Ibrahim M, Yar AM, Zaman G, Yan C, Khurshid M, Bokhari H (2018) Genome sequence and analysis of Mycobacterium tuberculosis strain SWLPK. J Glob Antimicrob Resist 13:211–213
Liu L, Feng Y, Tang G, Lin J, Huang W, Qiao F, Zong Z (2018) Carbapenem-resistant isolates of the Klebsiella pneumoniae complex in Western China: the common ST11 and the surprising hospital-specific types. Clin Infect Dis 67:S263–S265
Pires DP, Oliveira H, Melo LD, Sillankorva S, Azeredo J (2016) Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol 100:2141–2151
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858
Adriaenssens E, Brister JR (2017) How to name and classify your phage: an informal guide. Viruses. https://doi.org/10.3390/v9040070
Hsieh PF, Lin HH, Lin TL, Chen YY, Wang JT (2017) Two T7-like bacteriophages, K5-2 and K5-4, each encodes two capsule depolymerases: isolation and functional characterization. Sci Rep 7:4624
Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR (2019) Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539
Hung CH, Kuo CF, Wang CH, Wu CM, Tsao N (2011) Experimental phage therapy in treating Klebsiella pneumoniae-mediated liver abscesses and bacteremia in mice. Antimicrob Agents Chemother 55:1358–1365
Manohar P, Nachimuthu R, Lopes BS (2018) The therapeutic potential of bacteriophages targeting gram-negative bacteria using Galleria mellonella infection model. BMC Microbiol 18:97
El Haddad L, Harb CP, Gebara MA, Stibich MA, Chemaly RF (2019) A systematic and critical review of bacteriophage therapy against multidrug-resistant ESKAPE organisms in humans. Clin Infect Dis 69:167–178
Cao F, Wang X, Wang L, Li Z, Che J, Wang L, Li X, Cao Z, Zhang J, Jin L, Xu Y (2015) Evaluation of the efficacy of a bacteriophage in the treatment of pneumonia induced by multidrug resistance Klebsiella pneumoniae in mice. Biomed Res Int 2015:752930
Garneau JR, Depardieu F, Fortier LC, Bikard D, Monot M (2017) PhageTerm: a tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci Rep 7:8292
Philipson CW, Voegtly LJ, Lueder MR, Long KA, Rice GK, Frey KG, Biswas B, Cer RZ, Hamilton T, Bishop-Lilly KA (2018) Characterizing phage genomes for therapeutic applications. Viruses 10(4):188
Tzipilevich E, Habusha M, Ben-Yehuda S (2017) Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168(186–199):e112
Du Toit A (2019) Phage induction in different contexts. Nat Rev Microbiol 17:126–127
Born Y, Knecht LE, Eigenmann M, Bolliger M, Klumpp J, Fieseler L (2019) A major-capsid-protein-based multiplex PCR assay for rapid identification of selected virulent bacteriophage types. Arch Virol 164:819–830
Gigante AM, Hampton CM, Dillard RS, Gil F, Catalao MJ, Moniz-Pereira J, Wright ER, Pimentel M (2017) The Ms6 mycolyl-arabinogalactan esterase LysB is essential for an efficient mycobacteriophage-induced lysis. Viruses. https://doi.org/10.3390/v9110343
Bertozzi Silva J, Storms Z, Sauvageau D (2016) Host receptors for bacteriophage adsorption. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnw002
Manohar P, Tamhankar AJ, Lundborg CS, Nachimuthu R (2019) Therapeutic characterization and efficacy of bacteriophage cocktails infecting Escherichia coli, Klebsiella pneumoniae, and enterobacter species. Front Microbiol 10:574
Zhang W, Mi Z, Yin X, Fan H, An X, Zhang Z, Chen J, Tong Y (2013) Characterization of Enterococcus faecalis phage IME-EF1 and its endolysin. PLoS One 8:e80435
Khan A, Sharma D, Faheem M, Bisht D, Khan AU (2017) Proteomic analysis of a carbapenem-resistant Klebsiella pneumoniae strain in response to meropenem stress. J Glob Antimicrob Resist 8:172–178
Sharma D, Garg A, Kumar M, Khan AU (2019) Proteome profiling of carbapenem-resistant K. pneumoniae clinical isolate (NDM-4): exploring the mechanism of resistance and potential drug targets. J Proteomics 200:102–110
Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN (2004) SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629–1631
Jousset AB, Rosinski-Chupin I, Takissian J, Glaser P, Bonnin RA, Naas T (2018) Transcriptional landscape of a bla KPC-2 plasmid and response to imipenem exposure in Escherichia coli TOP10. Front Microbiol 9:2929
Acknowledgements
We would like to thank Dr. Supeng Yin (Fourth Medical Center of PLA General Hospital) for English language editing.
Funding
This work was funded by the National Natural Science Foundation of China (No. 81571896 and no. 81772073), the Foundation of State Key Laboratory of Trauma, Burns and Combined Injury (no. SKLZZ201708), and the Technological Innovation Plan in Major Fields of Southwest Hospital, Key Projects (no. SWH2016ZDCX2001).
Author information
Authors and Affiliations
Contributions
YZP and YLG conceived and designed the experiments. YLS, YC, ZCY, YLZ, XZL and YJC performed the experiments. PC and BY analyzed the data. MXL, CZ, XQL, ZQY and JC contributed reagents and materials. YLS wrote the paper. All authors contributed toward revising the paper and agreed to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Ethical approval
No human participants or animals were used by any of the authors in this study.
Additional information
Handling Editor: T. K. Frey.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
(A) Plaque morphology of kpssk3; (B) transmission electron micrograph of kpssk3 (TIFF 1384 kb)
Supplementary Fig. S1
(A) Plaque morphology of kpssk3; (B) transmission electron micrograph of kpssk3 (TIFF 235 kb)
Supplementary Fig. S3
(A) Thermal stability of kpssk3; (B) pH stability of kpssk3 (TIFF 179 kb)
Rights and permissions
About this article
Cite this article
Shi, Y., Chen, Y., Yang, Z. et al. Characterization and genome sequencing of a novel T7-like lytic phage, kpssk3, infecting carbapenem-resistant Klebsiella pneumoniae. Arch Virol 165, 97–104 (2020). https://doi.org/10.1007/s00705-019-04447-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04447-y