Abstract
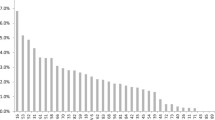
Recently, a clinical need for an improved human papilloma virus (HPV) test that covers a broad range of genotypes has emerged as a valuable primary screening tool for cervical lesions. The liquid bead microarray (LBMA) assay is a recently developed high-throughput platform covering a broad range of genotypes. Here, we compared the clinical performance of two recently developed LBMA assays, GeneFinderTM HPV Liquid Bead Microarray (GeneFinder) and CareGENETM HPV genotyping kit-O (CareGENE), in the Korean general population. A total of 3,148 cervical swabs were tested by the GeneFinder and CareGENE assays. Cases with discrepant results between the two assays were subjected to direct sequencing as a reference method for evaluating the performance of the two LBMA assays. Among all swabs tested, 12.6% showed HPV positivity, and the prevalent HPV genotypes were HPV53, 70, 16, 39, and 51, in that order. The concordance rates between the two assays for the detection of HPV and for genotyping were 96.6% (kappa = 0.836) and 94.5% (kappa = 0.779), respectively. The two LBMA assays showed comparable sensitivity and specificity for HPV detection (GeneFinder: sensitivity 94.4% and specificity 98.7%, CareGENE: sensitivity 89.8% and specificity 99.6%) and for genotyping (GeneFinder: sensitivity 91.0% and specificity 96.6%, CareGENE: sensitivity 90.2% and specificity 99.1%). This is the first demonstration that CareGENE has comparable clinical performance to GeneFinder, which has been established to show excellent performance for screening HPV in previous studies. Both LBMA platforms are thus considered to be valuable tools for HPV detection and genotyping to improve cervical screening in the general population.


Similar content being viewed by others
References
Wentzensen N, Vinokurova S, MvK Doeberitz (2004) Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res 64(11):3878–3884. https://doi.org/10.1158/0008-5472.Can-04-0009
Cuzick J, Szarewski A, Cubie H, Hulman G, Kitchener H, Luesley D, McGoogan E, Menon U, Terry G, Edwards R, Brooks C, Desai M, Gie C, Ho L, Jacobs I, Pickles C, Sasieni P (2003) Management of women who test positive for high-risk types of human papillomavirus: the HART study. Lancet (London, England) 362(9399):1871–1876. https://doi.org/10.1016/s0140-6736(03)14955-0
Poljak M, Kocjan BJ, Ostrbenk A, Seme K (2016) Commercially available molecular tests for human papillomaviruses (HPV): 2015 update. J Clin Virol 76(Suppl 1):S3. https://doi.org/10.1016/j.jcv.2015.10.023
Jun S-Y, Park ES, Kim J, Kang J, Lee JJ, Bae Y, Kim S-I, Maeng L-S (2015) Comparison of the Cobas 4800 HPV and HPV 9G DNA Chip tests for detection of high-risk human papillomavirus in cervical specimens of women with consecutive positive hpv tests but negative pap smears. PLoS One 10(10):e0140336. https://doi.org/10.1371/journal.pone.0140336
Castle PE, Sadorra M, Lau T, Aldrich C, Garcia FAR, Kornegay J (2009) Evaluation of a prototype real-time PCR assay for carcinogenic human papillomavirus (HPV) detection and simultaneous HPV genotype 16 (HPV16) and HPV18 genotyping. J Clin Microbiol 47(10):3344–3347. https://doi.org/10.1128/JCM.00725-09
Nayar R, Wilbur DC (2015) The pap test and Bethesda 2014. Acta Cytol 59(2):121–132
Halec G, Schmitt M, Dondog B, Sharkhuu E, Wentzensen N, Gheit T, Tommasino M, Kommoss F, Bosch FX, Franceschi S, Clifford G, Gissmann L, Pawlita M (2013) Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer. Int J Cancer 132(1):63–71. https://doi.org/10.1002/ijc.27605
Katki HA, Kinney WK, Fetterman B, Lorey T, Poitras NE, Cheung L, Demuth F, Schiffman M, Wacholder S, Castle PE (2011) Cervical cancer risk for women undergoing concurrent testing for human papillomavirus and cervical cytology: a population-based study in routine clinical practice. Lancet Oncol 12(7):663–672. https://doi.org/10.1016/S1470-2045(11)70145-0
Castle PE, Gravitt PE, Solomon D, Wheeler CM, Schiffman M (2008) Comparison of linear array and line blot assay for detection of human papillomavirus and diagnosis of cervical precancer and cancer in the atypical squamous cell of undetermined significance and low-grade squamous intraepithelial lesion triage study. J Clin Microbiol 46(1):109–117. https://doi.org/10.1128/jcm.01667-07
Dunn ST, Allen RA, Wang S, Walker J, Schiffman M (2007) DNA extraction: an understudied and important aspect of HPV genotyping using PCR-based methods. J Virol Methods 143(1):45–54. https://doi.org/10.1016/j.jviromet.2007.02.006
Ko K, Kwon MJ, Lee EH, Woo HY, Park H (2017) Comparison of GeneFinder human papillomavirus (HPV) liquid beads microarray PCR Kit and Hybrid capture 2 assay for detection of HPV Infection. J Clin Lab Anal 31:2. https://doi.org/10.1002/jcla.22025
Cho EJ, Do JH, Kim YS, Bae S, Ahn WS (2011) Evaluation of a liquid bead array system for high-risk human papillomavirus detection and genotyping in comparison with Hybrid Capture II, DNA chip and sequencing methods. J Med Microbiol 60(2):162–171. https://doi.org/10.1099/jmm.0.021642-0
Ozaki S, Kato K, Abe Y, Hara H, Kubota H, Kubushiro K, Kawahara E, Inoue M (2014) Analytical performance of newly developed multiplex human papillomavirus genotyping assay using Luminex xMAP™ technology (Mebgen™ HPV Kit). J Virol Methods 204:73–80. https://doi.org/10.1016/j.jviromet.2014.04.010
Feng Q, Cherne S, Winer RL, Balasubramanian A, Lee S-K, Hawes SE, Kiviat NB, Koutsky LA (2009) Development and evaluation of a liquid bead microarray assay for genotyping genital human papillomaviruses. J Clin Microbiol 47(3):547–553. https://doi.org/10.1128/JCM.01707-08
Jiang HL, Zhu HH, Zhou LF, Chen F, Chen Z (2006) Genotyping of human papillomavirus in cervical lesions by L1 consensus PCR and the Luminex xMAP system. J Med Microbiol 55(Pt 6):715–720. https://doi.org/10.1099/jmm.0.46493-0
Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T (2006) Bead-based multiplex genotyping of human papillomaviruses. J Clin Microbiol 44(2):504–512. https://doi.org/10.1128/jcm.44.2.504-512.2006
Jin MS, Lee H, Kim MA, Park IA, Lee C, An HJ, Shim B, Moon JH, Won JK, Ryu HS (2018) Novel cytomorphologic characteristics suggesting human papillomavirus infection in patients diagnosed as negative for intraepithelial lesion or malignancy and a comparison of diagnostic performance of three human papillomavirus tests. Diagn Cytopathol 46(10):833–839. https://doi.org/10.1002/dc.24049
Ryu HS, Park IA, Park SY, Jung YY, Park SH, Shin HC (2013) A pilot study evaluating liquid-based fine needle aspiration cytology of breast lesions: a cytomorphological comparison of SurePath(R) liquid-based preparations and conventional smears. Acta Cytol 57(4):391–399. https://doi.org/10.1159/000351306
Yoon YA, Kim BH, Heo SH, Kim HJ, Choi YJ (2018) Comparative evaluation of the Omniplex-HPV and RFMP HPV PapilloTyper for detecting human papillomavirus genotypes in cervical specimens. Adv Virol 163(4):969–976. https://doi.org/10.1007/s00705-017-3687-4
Hong G, Park YG, Jin HW, Choi SK, Chi HY, Cho WJ, Lee JH, Han S, Hong SP (2018) Comparison of the clinical performance of OmniPlex-HPV and GeneFinder HPV for the detection and genotyping of human papillomaviruses in cervical specimens. J Med Microbiol 67(9):1279–1286. https://doi.org/10.1099/jmm.0.000798
Choi J, Park Y, Lee EH, Kim S, Kim JH, Kim HS (2013) Detection and genotyping of human papillomavirus by five assays according to cytologic results. J Virol Methods 187(1):79–84. https://doi.org/10.1016/j.jviromet.2012.09.005
Bruni L, Diaz M, Castellsagué M, Ferrer E, Bosch FX, de Sanjosé S (2010) Cervical human papillomavirus prevalence in 5 continents: Meta-analysis of 1 million women with normal cytological findings. J Infect Dis 202(12):1789–1799. https://doi.org/10.1086/657321
de Sanjosé S, Diaz M, Castellsagué X, Clifford G, Bruni L, Muñoz N, Bosch FX (2007) Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: a meta-analysis. Lancet Infect Dis 7(7):453–459. https://doi.org/10.1016/S1473-3099(07)70158-5
Schmitt M, Depuydt C, Benoy I, Bogers J, Antoine J, Arbyn M, Pawlita M (2013) Prevalence and viral load of 51 genital human papillomavirus types and three subtypes. Int J Cancer 132(10):2395–2403. https://doi.org/10.1002/ijc.27891
Iftner T, Villa LL (2003) Chapter 12: human papillomavirus technologies. J Natl Cancer Inst Monogr 31:80–88
de Sanjose S, Quint WG, Alemany L, Geraets DT, Klaustermeier JE, Lloveras B, Tous S, Felix A, Bravo LE, Shin HR, Vallejos CS, de Ruiz PA, Lima MA, Guimera N, Clavero O, Alejo M, Llombart-Bosch A, Cheng-Yang C, Tatti SA, Kasamatsu E, Iljazovic E, Odida M, Prado R, Seoud M, Grce M, Usubutun A, Jain A, Suarez GA, Lombardi LE, Banjo A, Menendez C, Domingo EJ, Velasco J, Nessa A, Chichareon SC, Qiao YL, Lerma E, Garland SM, Sasagawa T, Ferrera A, Hammouda D, Mariani L, Pelayo A, Steiner I, Oliva E, Meijer CJ, Al-Jassar WF, Cruz E, Wright TC, Puras A, Llave CL, Tzardi M, Agorastos T, Garcia-Barriola V, Clavel C, Ordi J, Andujar M, Castellsague X, Sanchez GI, Nowakowski AM, Bornstein J, Munoz N, Bosch FX (2010) Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol 11(11):1048–1056. https://doi.org/10.1016/s1470-2045(10)70230-8
Agreda PM, Beitman GH, Gutierrez EC, Harris JM, Koch KR, LaViers WD, Leitch SV, Maus CE, McMillian RA, Nussbaumer WA, Palmer ML, Porter MJ, Richart GA, Schwab RJ, Vaughan LM (2013) Long-term stability of human genomic and human papillomavirus DNA stored in BD SurePath and Hologic PreservCyt liquid-based cytology media. J Clin Microbiol 51(8):2702–2706. https://doi.org/10.1128/jcm.00759-13
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose.
Additional information
Handling Editor: Carolina Scagnolari.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Moon, J.H., Jeong, K., Kim, K. et al. Comparison of clinical performance of two high-throughput liquid bead microarray assays, GeneFinder and CareGENE, for cervical screening in the general population. Arch Virol 164, 2699–2706 (2019). https://doi.org/10.1007/s00705-019-04379-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-019-04379-7