Abstract
A new family of viruses named Sphaerolipoviridae has been proposed recently. It comprises icosahedral, tailless haloarchaeal viruses with an internal lipid membrane located between the protein capsid and the dsDNA genome. The proposed family Sphaerolipoviridae was divided into two genera: Alphasphaerolipovirus, including Haloarcula hispanica viruses SH1, PH1 and HHIV-2, and Betasphaerolipovirus, including Natrinema virus SNJ1. Here, we propose to expand the family Sphaerolipoviridae to include a group of bacteriophages infecting extreme thermophilic Thermus thermophilus and sharing a number of structural and genomic properties with archaeal sphaerolipoviruses. This new group comprises two members, lytic phage P23-77 and temperate phage IN93, as well as putative members P23-72 and P23-65H. In addition, several related proviruses have been discovered as integrated elements in bacterial genomes of the families Thermus and Meiothermus. Morphology of the virus particles and the overall capsid architecture of these bacteriophages resembles that of archaeal members of the Sphaerolipoviridae, including an unusual capsid arrangement in a T = 28 dextro lattice. Alpha- and betasphaerolipoviruses share with P23-77-like bacteriophages a conserved block of core genes that encode a putative genome-packaging ATPase and the two major capsid proteins (MCPs). The recently determined X-ray structure of the small and large MCPs of P23-77 revealed a single beta-barrel (jelly-roll) fold that is superimposable with the cryo-EM density maps of the SH1 capsomers. Given the common features of these viruses, we propose to include the so far unclassified P23-77-like bacteriophages into a new genus, “Gammasphaerolipovirus”, within the family Sphaerolipoviridae.
Similar content being viewed by others
Introduction
Organisms of the two prokaryotic domains, Bacteria and Archaea, differ radically in terms of their associated virospheres. Viruses infecting archaea often display unique virion morphotypes that are not observed among bacteriophages. This is especially the case with hyperthermohilic archaeal viruses, which are currently classified into ten different families [47], none of which has relatives among bacteriophages [3]. The only viral group that currently bridges the bacterial and archaeal virospheres is the order Caudovirales (families Myoviridae, Siphoviridae and Podoviridae). Initially, it was proposed that the presence of tailed dsDNA viruses in halophilic and methanogenic archaea is a result of horizontal virus transfer from bacteria [46]. However, the subsequent identification of these viruses in a wide range of archaeal phyla [5, 26], including the deeply branching ones [28], favored an ancient association of the Caudovirales with Archaea. Recently, another group of archaeal viruses sharing features with bacteriophages has been described. This new viral group is the topic of the present communication.
In 2003, a spherical halovirus, SH1, infecting the euryarchaeon Haloarcula hispanica was isolated from Lake Serpentine in Western Australia [13] and characterized in detail during the next few years [7, 23, 29, 42–44]. The particle morphology and genome structure of this virus resemble those of bacteriophages belonging to the family Tectiviridae [2]. It has an icosahedral capsid with spikes on the five-fold vertices, an inner lipid membrane, and no tail. The linear dsDNA genome has inverted terminal repeats and proteins attached to the termini [7, 43]. However, closer analysis of the 31-kb genome and the capsid structure revealed that SH1 differs markedly from members of the Tectiviridae. In fact, the capsid geometry of SH1 differed from all other virus capsid types described at the time [23]. The capsid is arranged in a novel T = 28 lattice, which is skewed to the right (T = 28 dextro). The bulk of the capsid is constructed from two (one ‘small’ and one ‘large’) major capsid proteins (MCP), which are arranged into two distinct capsomer types. Unusual horn-like spikes emerge from the vertices of the icosahedral capsid. A lipid membrane is located between the capsid shell and the genome, with the lipids being selectively acquired from the host during particle assembly [7].
So far, two other lytic H. hispanica viruses – HHIV-2 [19] and PH1 [45] – have been described, both of them being highly similar to SH1 in terms of virus morphology, gene and protein homology, gene synteny, and genomic structure. Recently, the fourth SH1-like membrane-containing halovirus, Natrinema sp. J7-1 virus SNJ1, has been isolated [59]. Unlike other SH1-like viruses, SNJ1 has a temperate life cycle, and its genome is a circular, rather than linear, dsDNA molecule. Notably, the SNJ1 genome has been originally reported as a plasmid pHH205 [57]. The SNJ1 genome shares little similarity to the SH1-type viruses at the nucleotide sequence level, but a clear relatedness is found at the protein sequence level and in the arrangement of the viral core genes encoding the predicted genome packaging ATPase and the small and large MCPs. In addition, proviruses and elements related to the above-mentioned viruses had been identified in the genomes of several members of the Halobacteriaceae [21, 22, 45].
Due to distinctiveness of SH1-like viruses, a new viral family, Sphaerolipoviridae (from the Latin sphaero, for “sphere”, and the Greek lipos, for “fat”), has been proposed for their classification by Mike Dyall-Smith, Kate Porter and Sen-Lin Tang [15]. The new family consists of two genera: genus Alphasphaerolipovirus, comprising three species (SH1, HHIV-2 and PH1), and genus Betasphaerolipovirus, including one species (SNJ1). The proposal is currently under evaluation by the International Committee on Taxonomy of Viruses (ICTV).
More recently, a group of viruses infecting thermophilic bacteria of the family Thermaceae has been isolated and described. These bacteriophages, exemplified by Thermus thermophilus phage P23-77, display genomic and structural features that closely resemble those of archaeal sphaerolipoviruses. In this paper, we suggest the formation of a new genus in the family Sphaerolipoviridae, named “Gammasphaerolipovirus”, for classification of so far unclassified Thermus viruses. We summarize the available genomic and structural information on this viral group and examine their relatedness to the archaeal sphaerolipoviruses. The classification of bacterial and archaeal viruses within the same family establishes a new link between bacterial and archaeal virospheres.
Members of the novel family Sphaerolipoviridae
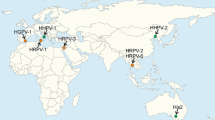
Members of this newly proposed family comprise icosahedral, nontailed dsDNA viruses with a round appearance and an internal lipid membrane (Fig. 1). The distinguishing features common to the members of this viral group include overall virion morphology, genome size and organization, gene synteny, sequence homology, and structure of the capsid. They differ from all other described viruses infecting Archaea, Bacteria or Eukarya. The general characteristics of members of the Sphaerolipoviridae are highlighted in Tables 1 and 2. All members of the Sphaerolipoviridae infect hosts from extreme environments, which are either halophilic archaea in the case of genera Alphasphaerolipovirus and Betasphaerolipovirus or extreme thermophilic bacteria of the genus Thermus in the case of the newly proposed genus Gammasphaerolipovirus. Members have a narrow host range, typically infecting just one or two strains of the same host species. Sphaerolipoviruses display global geographical distribution, with both halophilic and thermophilic members having been isolated from different continents (Table 2). They all share a conserved block of viral core genes: a gene for a putative genome packaging ATPase in close proximity to sequentially arranged genes encoding the two major capsid proteins.
(A) Three-dimensional image reconstruction of a P23-77 virion. Symmetry axes are designated with a black ellipse (2-fold), triangle (3-fold) and pentagon (5-fold). C, capsid shell; M, membrane; D, DNA. (B) Comparison of bacteriophage P23-77 and archaeal virus SH1. Surface representations of the two virions are viewed down a 3-fold symmetry axis. Symmetry axes are indicated with a white ellipse (2-fold), triangle (3-fold) and pentagon (5-fold). Capsomers of related symmetry are shown in the same color. Image reconstruction and surface representations are reproduced from ref. [20] with permission from Elsevier
Genus Alphasphaerolipovirus
The genus Alphasphaerolipovirus consists of three haloviruses, SH1, PH1 and HHIV-2. All three viruses have linear genomes of ~30 kb in size (Fig. 2A) with inverted terminal repeats and proteins attached to the genomic termini [7, 43], suggesting a protein-primed mechanism of genome replication. However, no corresponding DNA-polymerase-encoding genes are found in viral or host genomes, indicating that a different mode of replication might be employed. The three genomes are highly similar with respect to sequence homology and gene synteny. The overall identities between the nucleotide sequences of alphasphaerolipoviruses are as follows: 72 % between SH1 and PH1, 59 % between SH1 and HHIV-2, and 54 % between PH1 and HHIV-2. All three viruses infect the same host, Haloarcula hispanica. Peculiarly, however, HHIV-2 was isolated from a saltern in Margherita di Savoia, Italy [5], whereas SH1 and PH1 were both isolated in Western Australia from Serpentine Lake and Pink Lake, respectively [13, 45]. The higher level of sequence similarity between SH1 and PH1 may reflect the geographical proximity of the corresponding sampling points compared to that from which HHIV-2 has been isolated. Since SH1 was the first representative and most extensively studied member of the genus, it was chosen as the type member [15]. Alphasphaerolipoviruses display a narrow host range. Besides the common host, H. hispanica ATCC33960 [24], Halorubrum strain CSW 2.09.4 [42] was identified as an alternative host for SH1 and PH1, while SH1 and HHIV-2 could also infect Haloarcula sp. PV7 [5].
(A) Genomes of proposed alphasphaerolipoviruses SH1 (NC_007217), PH1 (KC252997) and HHIV-2 (JN968479) and (B) betasphaerolipovirus SNJ1 (NC_003158). The genome of SNJ1 is linearized for clearer presentation. ORFs are represented by arrows. Genes encoding core proteins are shown in red (ATPase) and dark blue (major capsid proteins, MCPs). Genes encoding structural components of the virions are in light blue (VP for virion protein, PB for protein band). ORFs, whose gene products showed significant similarity between the two genera, are marked in yellow (color figure online)
In addition, haloarchaea of five and three different genera, respectively, could be transfected with DNA of PH1 [45] and SH1 [43], demonstrating that the cell machinery of several halophilic organisms is suitable for virus propagation. Thus, the limited host range of alphasphaerolipoviruses is likely to reflect the failure of the viruses to adsorb to or to enter the host cell.
Genus Betasphaerolipovirus
The only member of the genus Betasphaerolipovirus is the temperate halovirus SNJ1, which was isolated from a salt mine in Hubei, China [52]. It could be induced from a halophilic euryarchaeon Natrinema sp. J7-1 and infects Natrinema sp. J7-2 [37], the only host identified for this virus so far. The sequence of the circular dsDNA is identical to the previously described Natrinema sp. J7-1 plasmid pHH205 [57, 59]. Virion size and morphology are shared with other members of the family (Table 1). The circular dsDNA genome is significantly smaller than the linear genomes of alphasphaerolipoviruses; however, it is in the size range of the circular genomes of the newly proposed gammasphaerolipoviruses (Table 1, also see below). SNJ1 shares several genes with the members of Alphasphaerolipovirus, including the ones for the two MCPs and the predicted genome packaging ATPase (Fig. 2B). Ten structural proteins have been identified in the virion of SNJ1 [59], which is in the same range as for alphasphaerolipoviruses.
Proviruses related to members of the genera Alphasphaerolipovirus and Betasphaerolipovirus
Several loci have been detected in the genomes of haloarchaea that appear to be of viral origin and show similarity at the nucleotide and protein levels to alpha- and betasphaerolipoviruses [19, 21, 22, 45]. They are likely to represent either functional or defective proviruses. The genomic loci identified in Haladaptatus paucihalophilus (designated as HalaPauP1 in Table 2) and Halobiforma lacisalsi (designated as HaloLacP1 in Table 2) are mosaics containing mixed assemblages of genes homologous to those of alphasphaerolipoviruses SH1/PH1 and HHIV-2 [45]. The putative provirus HalaPauP1 is ~17,026 bp in length and is found in the genome of Haladaptatus paucihalophilus DX253 (GenBank accession number NZ_AEMG01000027; ZOD2009_19038 to ZOD2009_19153). The putative provirus HaloLacP1 (~17,758 bp) stretches over genomic contigs 123 and 124 of Halobiforma lacisalsi AJ5 (GenBank accession numbers NZ_AGFZ01000123, NZ_AGFZ01000124) and is flanked by IS4 family transposase genes on both ends (locus tag HlacAJ_010100019767 on contig 123 and locus tag HlacAJ_010100019917 on contig 124).
The provirus HaloMukP1 present in the genome of Halomicrobium mukohataei DSM12286 [22], by contrast, is related to betasphaerolipovirus SNJ1. The ~15,814-bp HaloMukP1 sequence is part of a previously described ~40-kb locus of “virus and plasmid related elements” (ViPREHmuk1) [14, 45]. ViPREHmuk1 is flanked by a tRNA gene for alanine (tRNA-Ala) on one end and a partial copy of the identical tRNA-Ala gene on the other end. The H. mukohataei DSM12286 genome carries another ~19,058-bp provirus sequence, HaloMukP2, spanning the region between Hmuk_1699 and Hmuk_1732, followed by a tRNA gene for proline (tRNA-Pro). HaloMukP1 and HaloMukP2 are highly similar in terms of gene synteny and protein homology; particularly, the MCPs are nearly 100 % identical [22].
Another putative provirus, IHP (for integrated Haloarcula provirus; ~18,983 bp), is integrated into the genome of Haloarcula marismortui ATCC 43049 and is flanked by a tRNA-Pro gene on one side and an integrase gene on the other [21, 22]. IHP shares genes with both alphasphaerolipoviruses and SNJ1.
In conclusion, proviruses related to alpha- and betasphaerolipoviruses are integrated into the genomes of diverse haloarchaea, where they are typically found adjacent to tRNA genes, a common target site for the integration of various temperate viruses and plasmids.
The new genus Gammasphaerolipovirus
Over the last few years, a number of bacteriophages infecting extreme thermophilic bacteria of the genus Thermus have been isolated, and this group includes four representatives: P23-77, P23-72, P23-65H [58] and IN93 [32]. Following detailed characterization, it became apparent that these bacterial viruses display striking similarity to archaeal sphaerolipoviruses (as detailed below), rather than to any other group of bacteriophages. Thus, to acknowledge this relationship, we propose the creation of a new genus, “Gammasphaerolipovirus” within the family Sphaerolipoviridae.
The most extensively studied virus of this group, P23-77, was isolated from an alkaline hot spring on the North Island of New Zealand, together with bacteriophages P23-72 and P23-65H [58]. The sampling expedition focusing on isolation of Thermus viruses for potential biotechnological applications was directed by the Promega Corporation [58]. These viruses were originally designated as tectiviruses due to the overall morphology of the virus particles. All three viruses were found to infect the same host and had practically identical growth cycles and virus particle yields. They also showed very similar protein patterns in SDS-PAGE analysis [20]. Due to its superior stability, P23-77 was selected for more detailed characterization. Although there are no genome sequence data available for P23-72 and P23-65H, the observed similarity in virion morphology and composition indicates that the two viruses belong in the same genus with P23-77.
IN93 was isolated from hot-spring soil in Japan [32]. It is a temperate phage that could be induced from lysogenic host Thermus thermophilus TZ2. Like alpha- and betasphaerolipoviruses, gammasphaerolipoviruses have a narrow host range. P23-77, P23-72 and P23-65H share a common host, T. thermophilus ATCC33923. In addition, P23-77 and P23-72 have been shown to form plaques on T. thermophilus ATCC27978 [58]. IN93 replicates in T. thermophilus TZ2 and T. thermophilus HB8 [34].
We propose to designate phage P23-77 as the type member of the suggested genus on the basis of comprehensive analysis of its genome, capsid architecture, and high-resolution structure of capsid proteins (see below).
Morphology of gammasphaerolipoviruses
P23-77 virus particles are spherical and tailless and have an average diameter of 78 nm. Approximately 15-nm-long stick-like spikes emerge from the five-fold vertices ([20], Fig. 1A). An inner lipid membrane is located between the 6-nm-thick capsid and the circular dsDNA genome. The capsid and membrane are connected by proteins at the five-fold vertices. The lipids are selectively acquired from the host cell during virus assembly [21]. Neutral lipids and phosphoglycolipids were identified in the membrane of P23-77, while glycolipids, a substantial component of the host lipid composition, were almost completely excluded. The same is true for SH1, where glycolipids, which constitute approximately 20 % of the total host lipids, failed to be detected in viral lipid extracts [7]. Selective incorporation of lipids has been shown for other non-tailed, membrane-containing icosahedral viruses [8, 9, 30, 31], whereas the lipid composition of archaeal and bacterial pleomorphic viruses reflects the host lipid composition [4, 41]. Three of the 10 identified structural proteins of P23-77 have been shown to be associated with the capsid, and five with the membrane, while the remaining two could not be unambiguously located [21]. In contrast, of the 11 structural proteins identified in SH1, eight are capsid proteins and three are associated with the membrane [29]. In bacteriophage PRD1, the type member of the family Tectiviridae, most of the membrane-associated proteins are involved in virus entry [16]. The difference in the number of membrane proteins in P23-77 and SH1 may reflect differences in host entry – a reasonable assumption given that the two viruses infect hosts from two different domains of life.
The geometrical arrangement of capsomers in an icosahedral capsid can be defined by the triangulation (T) number [11]. The P23-77 capsid consists of 270 hexameric and 12 pentameric capsomers, arranged in a T = 28, dextro lattice. The only other characterized virus with such unusual capsid architecture is SH1 (Fig. 1B). This observation suggests that the two groups of viruses, one bacterial and the other archaeal, share a common ancestor.
Genome analysis
P23-77 and IN93 have circular dsDNA genomes. Their genome sizes are 17,036 bp with 37 assigned ORFs for P23-77 and 19,604 bp with 39 assigned ORFs for IN93 [21, 34]. We have assigned four additional ORFs (ORF40-43) to the genome of IN93 based on sequence comparisons (Fig. 3A, Online Resource 1). Most of the P23-77 genes have homologs in IN93, yet 78 % of the gene products lack similarity to any other protein sequences in public databases. The viral core proteins – the putative genome packaging ATPase (ORF13) and the small (ORF16) and large (ORF17) MCPs – are among the most conserved proteins in P23-77 and IN93, with sequence identities of 79, 74 and 79 %, respectively. The gene order is highly conserved in the two genomes. The main difference is the presence of an integration cassette encoding a LexA-like repressor, an endonuclease and an integrase required for the lysogenic cycle (ORFs 36-39, Fig. 3) in the genome of IN93. The genes of the integration cassette are the only ones located on the opposite strand with respect to the rest of the genes. P23-77 lacks the integration cassette, which is reflected by its smaller genome size and strictly lytic life style. A partial Thermus tRNA gene for isoleucine (tRNA-Ile) is located directly downstream of the integrase gene of IN93, enabling genome integration through recombination with an identical tRNA-Ile gene sequence in the host genome [34].
(A) Genomes of proposed gammasphaerolipoviruses P23-77 (NC_013197) and IN93 (NC_004462) and (B) related proviruses: TP1, identified in the genome of Thermus sp. RLM (NZ_AIJQ01000002.1), MeioSilP1, identified in the genome of Meiothermus silvanus DSM 9946 (NC_014212.1), and MeioRubP1 and P2, identified in the genome of Meiothermus ruber DSM 1279 (NC_013946.1). Genomes of P23-77 and IN93 are linearized for clearer presentation. ORFs are represented by arrows. ORF numbers are according to GenBank entry (P23-77, IN93) or position in the host genome (TP1, MeioSilP1, MeioRubP1and P2). Transcriptional units are indicated in brackets below the IN93 genome. Genes are shown in color if their gene products have been identified as structural components of the virus (VP = virion protein) or if a function is assigned according to experimental data or hits in BLAST searches. Genes of unknown function are colored black (shared by all members), grey (shared by at least two members) or white (found in only one member). Genes encoding viral core proteins, ATPase, and the two major capsid proteins (MCP), are marked in italics and bold-framed arrows
Besides the small and large MCPs, eight additional structural proteins have been identified in P23-77 (assigned as VP for virion protein in Fig. 3A). One is a minor capsid protein encoded by ORF11, which has no homologues in public databases or in related viruses or viral elements. Five are membrane-associated proteins (corresponding ORFs 15-17, 19, 20, 22 and 23). Whether VP29 (ORF29) is located in the capsid or membrane is unclear [21]. The homologous gene in IN93 (ORF23) encodes a functional lysozyme, which displays no sequence similarity to other known lysozymes. It acts specifically on Thermus species and is highly thermostable [33]. In the P23-77 genome, the thermophilic lysozyme-encoding gene is grouped together with genes coding for a putative transglycosylase (ORF31) and a protein with a WD40-like beta-propeller domain (ORF28; Fig. 3A). A beta-propeller head is found in the structure of PRD1 receptor binding protein P2 [56], implying a role in receptor recognition. A similar set of genes is found in the genomes of the related proviruses TP1 and MeioSilP1 (Fig. 3B). This block of genes may play a role in receptor recognition and host cell entry.
Transcription of viral genes has been experimentally investigated in the case of IN93; its genome was found to be divided into four transcriptional units and transcribed by the host RNA polymerase [34]. The first unit contains genes for DNA-binding and genome replication proteins (ORF28-ORF4, Fig. 3A). The second one includes genes for DNA-packaging and lytic enzymes (ORF5-ORF11). The third group of genes encodes the majority of structural proteins and proteins involved in virus entry (ORF12-ORF27). The fourth transcriptional unit (ORF36-39) is transcribed in the opposite direction when compared to units 1-3 and contains genes for proteins required for integration into the host genome. Transcriptional units 1 and 4 are likely to be transcribed during early stages of infection from Thermus promoters, whereas units 2 and 3 are transcribed later on from inherent IN93 promoters. Since the gene order is highly conserved in IN93 and P23-77, we assume similar transcriptional units for P23-77. Gene synteny is highest in the area where structural genes are located (transcriptional unit 3) and most variable in the area containing genes for DNA replication (unit 1). Transcriptional unit 2 (DNA-packaging and virus exit) also shows a variable gene content, which may reflect adaptation to the host. P23-77 has two lytic enzymes in this area, probably attributable to the strictly lytic life cycle. In the same region, IN93 carries a gene encoding a metallopeptidase, which was most likely acquired from the host.
Genome replication
As in the case of other members of the Sphaerolipoviridae, P23-77-like viruses do not encode their own DNA polymerases for genome replication. The ORF1 products of P23-77 and IN93 show 26 % identity to the replication initiation protein RepA of Thermus sp. plasmid pMY1 [12] for which a theta-type mechanism of replication was proposed. The IN93 ORF1 gene was used to create an Escherichia coli/Thermus thermophilus shuttle vector [35]. The plasmid successfully replicated in T. thermophilius, demonstrating that ORF1 indeed encodes the replication protein of IN93 and that it is sufficient for replication of circular dsDNA molecules. Thus, gammasphaerolipoviruses are replicated the same way as plasmids of Thermus species, most likely by a theta-type mode of replication.
Life cycle
Members of the proposed genus Gammasphaerolipovirus exhibit different life cycles (Table 1). IN93 is a temperate phage, which integrates into a tRNA gene in the host chromosome. The only other temperate sphaerolipovirus, SNJ1, persists within the cell in an extrachromosomal, episomal form [59]. P23-77, P23-72 and P23-65H are strictly lytic, with cell lysis occurring ~90 min postinfection [20]. The widespread occurrence of proviruses related to members of the Sphaerolipoviridae indicates that a temperate lifestyle is a preferred strategy for these viruses. Genome integration may increase the chances of survival in extreme environments, where cell growth rates are often low and virions are exposed to harsh conditions during transmission.
Proviruses related to gammasphaerolipoviruses
Several P23-77-like proviruses have been described [22]. Recently, we have identified a new proviral member, TP1 (for Thermus provirus 1), of the gammasphaerolipovirus group integrated into the genome of Thermus sp. RLM (isolated from a hot spring in Manikaran, India, NZ_AIJQ01000002). The putative provirus is integrated into a tRNA-Ile gene and spans a genomic region of 19,265 bp between genes RLTM_01505 and RLTM_01670 (Table 1 and Online Resource 1). Compared to the IN93 and P23-77 genomes, TP1 contains four additional ORFs (ORF8, 10, 33 and 34). Viral ORFs were designated according to their position in the bacterial genome, starting with ORF1 (RLTM_01505) to ORF38 (RLTM_01670) (Fig. 3B). Comparative genomic analysis shows that the TP1 genome is a mosaic and contains homologues of both IN93 and P23-77. The gene order is highly preserved with respect to P23-77/IN93. TP1 seems to contain all genes required for the production of virions, yet it remains to be elucidated if it could be induced from its host.
On the contrary, none of the other gammasphaerolipovirus-related sequences, which have been detected in the genomes of Meiothermus species [22], appears to represent a complete virus genome. The ~21,110-bp Meiothermus silvanus DSM9946 provirus MeioSilP1 contains an insertion of transposases and other provirus-unrelated genes in the integration cassette (ORF38–ORF44). Within the variable region for DNA binding and replication (transcriptional unit 1), the proviral genes are replaced by a set of genes apparently acquired from a bacterial genome (ORF25-ORF27). This sequence rearrangement explains the larger genome size of MeioSilP1 compared to P23-77 and IN93 and may be a consequence of host defense, rendering the virus defective [22].
In the case of Meiothermus ruber DSM1279 provirus MeioRubP1 [22], a set of genes proposed to be involved in host entry (ORF28–ORF30 in P23-77 and ORF22–ORF25 in IN93) is lacking, and this explains the smaller genome size of ~16,019 bp. Recently, we have detected remnants of another provirus in the genome of Meiothermus ruber DSM1279, MeioRubP2, which is nearly 100% identical to the sequence of MeioRubP1 but comprises only ORF1 to ORF23 (Fig. 3B, Online Resource 1). The ~10,430-bp MeioRubP2 sequence ranges from locus tag Mrub_0077 (ORF1, transglycosylase) to Mrub_0096 (ORF23) of the M. ruber genome (NC_013946.1). A homolog of MeioRubP1, ORF16, is missing in MeioRubP2.
All proviruses – except for MeioRubP2 – carry an IN93-related integration cassette (containing genes for a LexA-like repressor, an endonuclease and a phage integrase) and appear to be integrated into host tRNA genes (MeioRubP1, MeioRubP2 and MeioSilP1 into tRNA-Arg genes, and TP1 and IN93 into tRNA-Ile genes). Presumably, genome integration of the proviruses follows the same mechanism as described for IN93 [34]. The gene arrangement of the Thermus proviruses TP1, MeioSilP1 and MeioRubP1 – an integrase gene on one end of the provirus and a tRNA-gene directly following the last provirus-related gene on the opposite end of the sequence – is similar to the arrangement found in alpha-/betasphaerolipovirus-related proviruses IHP, HaloMukP1 and HaloMukP2 (see above). Integration into tRNA genes is common among bacterial and archaeal viruses [10, 48, 55] and allows multiple integrations into a single host genome. For example, six different integrated elements have been described in the genome of the euryarchaeon Methanococcus maripaludis C6 [27]. Identification of a proviruses related to bacterial and archaeal sphaerolipoviruses not only provided valuable data for comparative genomic analysis but also enabled a better resolution of the phylogenetic structure of the family Sphaerolipoviridae (see below).
Structure of major capsid proteins and capsid organization
Recently, the small (VP16) and the large (VP17) MCPs of P23-77 were crystallized and their structures determined [49, 50]. The core fold of both proteins is a nearly identical eight-stranded beta-barrel, which shows clear homology to the double beta-barrel capsid proteins of the structure-based PRD1/Adenovirus viral lineage (Fig. 4A-D). Members of the lineage infect hosts from all three domains of life [6, 25]. It has been suggested that P23-77 represents the earliest branch in the so-called vertical beta-barrel superlineage consisting of dsDNA viruses that use capsid proteins with a conserved beta-barrel core fold for capsid assembly and homologous genome-packaging ATPases [25, 50].
Structures of (A) P23-77 major capsid protein (MCP) VP17 (green, PDB ID code 3ZMN) and (B) P23-77 MCP VP16 (orange, PDB ID code 3ZMO) showing the eight-stranded single beta-barrel core fold. In addition, VP17 has an upper domain. VP16 exists as strand-swapped homodimer with each subunit completed by a strand from the other subunit (strand shown in grey). (C) VP16 superimposed on the lower domain of VP17. (D) P2 (yellow, PDB ID code 2VVF), the MCP of marine bacteriophage PM2, superimposed on the lower domain of VP17. PM2 is a member of the PRD1/adenovirus lineage. (E) P23-77 capsid protein structures fitted into the P23-77 virion cryo-EM reconstruction (EMDB ID code: emdb_1525). The upper domains of VP17 form turrets protruding from the capsomer base (inset). Molecular graphics and analyses were performed with the programs CCP4 Molecular Graphics Program v. 2.6.2 (36), PyMol (The PyMOL Molecular Graphics System, Version 1.5.0.5, Schrödinger, LLC, (http://www.pymol.org), and UCSF Chimera (40) (color figure online)
The capsid surface of P23-77 is covered with small turret-like protrusions [20]. The high-resolution structures of the P23-77 capsid proteins [50], fitted into the electron cryo-microscopy reconstruction (cryo-EM) of the P23-77 virion [20], showed that turrets are formed by the upper domain of VP17, while VP16 and the lower domain of VP17 form the base of the capsomers (Fig. 4E).
P23-77 has two distinct types of pseudohexameric capsomers (Fig. 5A). Both have two turrets, arranged either on the same side or on the opposite corners of the capsomer. Two types of turreted capsomers are also found in the alphaspaerolipovirus SH1, but they have either two or three turret protrusions (Fig. 5B) [23]. The model of the P23-77 capsid revealed an unexpected organization, which clearly differs from those in other members of the PRD1/adenovirus lineage [50]. The capsomers of PRD1/adenovirus lineage members consist of compact homotrimers, while in P23-77 the capsid building blocks are represented by several types of heteromultimers, with VP16 forming strong dimers across capsomer borders. The dimerization is clearly seen in the P23-77 virion cryo-EM reconstruction, where there are bridges of electron density between capsomers at the sites occupied by VP16 dimers (Fig. 5C). The same bridging pattern is observed in the cryo-EM reconstruction of the SH1 virion (Fig. 5D) [23]. Furthermore, the two coat proteins of SH1 are likely to participate in building the capsomers in the same way as in P23-77: small and large MCPs form the hexagonal base of the capsomer with turrets produced by an upper domain of the large MCP, and the strongest protein-protein interactions are between dimerized subunits that span adjacent capsomers. Indeed, the X-ray structures of the P23-77 MCPs are superimposable within the cryo-EM density maps of the SH1 capsomers, producing a reasonable fit [50]. Collectively, the same capsid geometry (T = 28), sequence and structural similarity between the corresponding MCPs as well as similar capsid stabilization principles utilized by P23-77-like and SH1-like viruses strongly suggest that the two viral groups have evolved from a common ancestor.
Comparison of the two capsomer types of P23-77 (A) and SH1 (B). The overall topology of the virions is similar, and both P23-77 (C) and SH1 (D) have bridges of electron density between adjacent capsomers (arrows). In P23-77, the bridges correspond to VP16 dimers that span across capsomer borders. EMDB ID codes for the cryo-EM maps used in producing the images: emdb_1350 (SH1) and emdb_1525 (P23-77). Molecular graphics and analyses were performed with the UCSF Chimera package (40)
Links to other viruses
The genomes of gammasphaerolipoviruses and related proviruses carry several genes that have homologues in other, unrelated viruses (Fig. 3). For example, homologues of the genes with predicted function in host recognition and cell entry (ORF28–ORF31 in P23-77 and ORF22–ORF26 in IN93) are found in the genomes of unrelated tailed siphoviruses P23-45 and P74-26, which also infect T. thermophilus [38]. Similarly, IN93 ORF24 and ORF43 despite having no relatives within P23-77 and related proviruses are also present in the same two siphoviruses. This block of entry-related genes is highly conserved and may be subject to horizontal gene transfer between unrelated Thermus phages that occupy the same ecological niche. Notably, in tectiviruses, genes involved in host recognition and cell entry were found to evolve more rapidly than those encoding other functions [51]. The presence of a conserved block of genes involved in virus entry in various, even unrelated Thermus phages implies a similar entry mechanism for these viruses. In contrast, HHIV-2, which infects the same host as SH1 and PH1, differs from the two viruses mainly in the putative spike-encoding region [19].
The products of IN93 ORF9 and provirus TP1 ORF21, located in the variable gene region, show similarity to the gene product VP2 of hyperthermophilic archaeal Sulfolobus and Acidianus spindle-shaped fuselloviruses. The presence of a gene related to archaeal viruses in the genomes of Thermus bacteriophages shows that viruses of extremophilic bacteria and archaea can share a common gene pool. TP1 encodes another protein (gene product of ORF1) that is similar to protein YS40_146, a protein of unknown function with three predicted transmembrane domains, encoded by myovirus phiYS40 infecting T. thermophilus HB8 [39].
In the genomic region encoding nonstructural proteins, P23-77 and TP1 carry a gene coding for a putative phosphoadenosine phosphosulfate reductase (PAPS-reductase) that is widespread in bacterial genomes and probably represents a moron [17]. Interestingly, a similar PAPS-reductase gene is found in the genome of bacteriophage SSIP-1, infecting halophilic bacterium Salisaeta sp. SP9-1 [1]. SSIP-1 is an icosahedral, tailless, inner-membrane-containing bacteriophage whose 44-kb genome shares no sequence similarity with sphaerolipoviruses. On the other hand, the virus encodes a PRD1-type genome-packaging ATPase and two major capsid proteins of nearly identical size that are arranged in the genome in a similar manner to the ATPase and MCP genes of members of the Sphaerolipoviridae. In addition, the capsid is formed by two types of capsomers. Consequently, SSIP-1 might be a highly divergent yet genuine member of the ancient virus group, which gave rise to bacterial and archaeal sphaerolipoviruses. However, further structural studies are needed to verify this hypothesis.
To conclude, in addition to the core of signature genes exclusive to gammasphaerolipoviruses, different members of the group carry a variable portion of genes that were horizontally acquired from various sources, including unrelated bacterial and archaeal viruses and plasmids.
Phylogenetic relationships within the Sphaerolipoviridae
All members of the Sphaerolipoviridae share a block of genes encoding a predicted packaging ATPase and the two MCPs (Figs. 2, 3). The ATPase gene is located in close proximity to the sequentially arranged MCP genes. In the betasphaerolipovirus SNJ1 and the proviruses HaloMukP1, HaloMukP2 and IHP as well as in gammasphaerolipoviruses and related proviruses, the ATPase gene is located one or two ORFs upstream of the MCP genes and transcribed in same direction. In alphaspaerolipoviruses and the related proviruses HalaPauP1 and HaloLacP1, the ATPase gene is separated by six genes from the sequentially arranged MCP genes and transcribed in the opposite direction. The ATPases of members of the Sphaerolipoviridae in addition to the classical Walker A and B motifs carry a signature motif common to all icosahedral dsDNA viruses with an internal membrane [18, 53] and initially identified in the packaging ATPase P9 of PRD1 (Online Resource 2, Fig. 3) [53], providing additional phylogenetic link between the Sphaerolipoviridae and the PRD1/adenovirus lineage.
Sequences of the core proteins are highly conserved between members of the same genus, with more than 70 % (small MCP) or 80 % (large MCP, ATPase) identity. The highest similarity is found between the core proteins of the alphashaerolipoviruses SH1 and PH1, which are more than 92 % identical. Although the overall similarity is low between genera, distinctive conserved residues could be identified in corresponding proteins, suggesting their relatedness (Online Resource 2, Figs. 1, 2, 3).
The phylogenetic relationship between the members of the Sphaerolipoviridae is illustrated in Fig. 6. Phylogenetic reconstruction based on the core gene products (small and large MCPs and the packaging ATPase) produced congruent trees, with the members of the proposed genera Alphasphaerolipovirus, Betasphaerolipovirus and Gammasphaerolipovirus falling into three distinct, well-supported clades. Based on the analysis of all three core proteins, the haloarchaeal proviruses IHP, HaloMukP1 and HaloMukP2 are related to the betasphaerolipovirus SNJ1, whereas HalaPauP1 and HaloLacP1 are clearly related to alphasphaerolipoviruses. As expected, proviruses identified in the Thermaceae genomes form a monophyletic clade with bacteriophages P23-77 and IN93. Notably, the phylogenetic grouping is consistent with the arrangement of the core genes in alpha- and betasphaerolipoviruses and the corresponding proviruses (see above). Thus, core proteins may serve as signature sequences for the identification of putative new members of the three genera in future data bank searches.
Molecular phylogenetic analysis of (A) large and (B) small major capsid protein and (C) ATPase sequences. Members that are confirmed to form virions are underlined. The evolutionary history was inferred by using the maximum-likelihood method based on the JTT amino acid substitution model. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. The percentage of trees in which the associated taxa clustered together is shown next to the branches. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5 (54)
Conclusion
The genomic and structural analyses presented above clearly indicate that P23-77-like viruses form a distinct group in the newly proposed family Sphaerolipoviridae. P23-77-like viruses infect thermophilic bacteria, whereas the other members of the family replicate in halophilic archaeal hosts. We suggest the name “Gammasphaerolipovirus” for this new genus in order to be consistent with the nomenclature proposed for the other two genera – “Alphasphaerolipovirus” and “Betasphaerolipovirus”. The proposal for the creation of the new genus Gammasphaerolipovirus is currently under consideration by the ICTV.
The relatedness of bacterial and archaeal viruses indicates, once again, that although hosts belong to different domains of life, their viruses can share common ancestors. Moreover, based on the major capsid protein structure of P23-77, the homologous ATPases, and the general virion organization with the inner membranes, Sphaerolipoviridae may represent a potentially primordial branch in the previously described structural lineage of dsDNA viruses that build their virions using the double beta-barrel capsid protein. The members of this lineage exhibit enormous variety in the characteristics of individual viruses [25]. The existence of Sphaerolipoviridae members with single beta-barrel capsid proteins pushes the border even further by illuminating the increasing variability in the ways these types of capsid proteins are utilized to assemble the virion shell. Overall, the vertical beta-barrel superlineage now comprises more than ten distinct virus families. The fact that single-beta-barrel viruses are present in two distinct domains of life may imply that their ancestor already existed before the separation of cellular organisms into the now established domains Bacteria and Archaea. An intriguing question for the future is whether dsDNA viruses with vertical single-beta-barrel capsid proteins are also present in the domain Eukarya, as in the case of their siblings with double-beta-barrel MCPs.
So far, the members of the Sphaerolipoviridae are restricted to extremophilic hosts. Given the low sequence homology between genera, it may be difficult to recognize potential mesophilic members using only sequence-based approaches. However, it would be interesting to know whether Sphaerolipoviridae is yet another unique group of viruses of extremophiles, or if similar viruses also exist in more moderate environments.
References
Aalto AP, Bitto D, Ravantti JJ, Bamford DH, Huiskonen JT, Oksanen HM (2012) Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1. Proc Natl Acad Sci USA 109:7079–7084
Abrescia NG, Cockburn JJ, Grimes JM, Sutton GC, Diprose JM, Butcher SJ, Fuller SD, San Martin C, Burnett RM, Stuart DI, Bamford DH, Bamford JK (2004) Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68–74
Ackermann HW, Prangishvili D (2012) Prokaryote viruses studied by electron microscopy. Arch Virol 157:1843–1849
Al-Shammar AJ, Smith PF (1981) Lipid composition of two mycoplasmaviruses, MV-Lg-L172 and MVL2. J Gen Virol 54:455–458
Atanasova NS, Roine E, Oren A, Bamford DH, Oksanen HM (2012) Global network of specific virus–host interactions in hypersaline environments. Environ Microbiol 14:426–440
Bamford DH, Burnet RM, Stuart DI (2002) Evolution of viral structure. Theor Popul Biol 61:461–470
Bamford DH, Ravantti JJ, Rönnholm G, Laurinavičius S, Kukkaro P, Dyall-Smith M, Somerharju P, Kalkkinen N, Bamford JK (2005) Constituents of SH1, a novel lipid-containing virus infecting the halophilic euryarchaeon Haloarcula hispanica. J Virol 79:9097–9107
Braunstein SN, Franklin RM (1971) Structure and synthesis of a lipid-containing bacteriophage: V phospholipids of the host BAL-31 and of the bacteriophage PM2. Virology 43:685–695
Brewer GJ, Goto RM (1983) Accessibility of phosphatidylethanolamine in bacteriophage PM2 and in its gram-negative host. J Virol 48:774–778
Canchaya C, Fournous G, Brüssow H (2004) The impact of prophages on bacterial chromosomes. Mol Microbiol 53:9–18
Caspar DT, Klug A (1962) Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol 27:1–24
de Grado M, Lasa I, Berenguer J (1998) Characterization of a plasmid replicative origin from an extreme thermophile. FEMS Microbiol Lett 165:51–57
Dyall-Smith M, Tang SL, Bath C (2003) Haloarchaeal viruses: how diverse are they? Res Microbiol 154:309–313
Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D (2011) Haloquadratum walsbyi: limited diversity in a global pond. PLoS One 6:e20968
Dyall-Smith M, Porter K, Tang SL (2013) Official website of the international committee on taxonomy of viruses (ICTV). (http://talk.ictvonline.org/files/proposals/taxonomy_proposals_prokaryote1/m/bact01/4633.aspx)
Grahn AM, Daugelavičius R, Bamford DH (2002) Sequential model of phage PRD1 DNA delivery: active involvement of the viral membrane. Mol Microbiol 46:1199–1209
Hendrix RW, Lawrence JG, Hatfull GF, Casjens S (2000) The origins and ongoing evolution of viruses. Trends Microbiol 8:504–508
Iyer LM, Makarova KS, Koonin EV, Aravind L (2004) Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res 32:5260–5279
Jaakkola ST, Penttinen RK, Vilén ST, Jalasvuori M, Rönnholm G, Bamford JK, Oksanen HM (2012) Closely related archaeal Haloarcula hispanica icosahedral viruses HHIV-2 and SH1 have nonhomologous genes encoding host recognition functions. J Virol 86:4734–4742
Jaatinen ST, Happonen LJ, Laurinmäki P, Butcher SJ, Bamford DH (2008) Biochemical and structural characterisation of membrane-containing icosahedral dsDNA bacteriophages infecting thermophilic Thermus thermophilus. Virology 379:10–19
Jalasvuori M, Jaatinen ST, Laurinavičius S, Ahola-Iivarinen E, Kalkkinen N, Bamford DH, Bamford JK (2009) The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J Virol 83:9388–9397
Jalasvuori M, Pawlowski A, Bamford JK (2010) A unique group of virus-related, genome-integrating elements found solely in the bacterial family Thermaceae and the archaeal family Halobacteriaceae. J Bact 192:3231–3234
Jäälinoja HT, Roine E, Laurinmäki P, Kivelä HM, Bamford DH, Butcher SJ (2008) Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc Natl Acad Sci USA 105:8008–8013
Juez G, Rodriguez-Valera F, Ventosa A, Kushner DJ (1986) Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec, nov., two new species of extremely halophilic archaebacteria. Syst Appl Microbiol 8:75–79
Krupovic M, Bamford DH (2008) Virus evolution: how far does the double β-barrel viral lineage extend? Nat Rev Microbiol 6:941–948
Krupovic M, Forterre P, Bamford DH (2010) Comparative analysis of the mosaic genomes of tailed archaeal viruses and proviruses suggests common themes for virion architecture and assembly with tailed viruses of bacteria. J Mol Biol 397:144–160
Krupovic M, Gribaldo S, Bamford DH, Forterre P (2010) The evolutionary history of archaeal MCM helicases: a case study of vertical evolution combined with hitchhiking of mobile genetic elements. Mol Biol Evol 27:2716–2732
Krupovic M, Spang A, Gribaldo S, Forterre P, Schleper C (2011) A thaumarchaeal provirus testifies for an ancient association of tailed viruses with archaea. Biochem Soc Trans 39:82–88
Kivelä HM, Roine E, Kukkaro P, Laurinavičius S, Somerharju P, Bamford DH (2006) Quantitative dissociation of archaeal virus SH1 reveals distinct capsid proteins and a lipid core. Virology 356:4–11
Laurinavičius S, Käkelä R, Somerharju P, Bamford DH (2004) Phospholipid molecular species profiles of tectiviruses infecting Gram-negative and Gram-positive hosts. Virology 322:328–336
Maaty WSA, Ortmann AC, Dlakič M, Schulstad K, Hilmer JK, Liepold L, Weidenheft B, Khayat R, Douglas T, Young MJ, Bothner B (2006) Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life. J Virol 80:7625–7635
Matsushita I, Yamashita N, Yokota A (1995) Isolation and characterization of bacteriophage induced from a new isolate of Thermus aquaticus. Microbiol Cult Collect 11:133–138
Matsushita I, Yanase H (2008) A novel thermophilic lysozyme from bacteriophage φIN93. Biochem Bioph Res Co 377:89–92
Matsushita I, Yanase H (2009) The genomic structure of Thermus bacteriophage ϕIN93. J Biochem 146:775–785
Matsushita I, Yanase H (2011) Characterization of the protein gp1 from bacteriophage φIN93. Biol Lett 48:47–56
McNicholas S, Potterton E, Wilson KS, Noble ME (2011) Presenting your structures: the CCP4mg molecular-graphics software. Acta Cryst D 67:386–394
Mei Y, Chen J, Sun D, Chen D, Yang Y, Shen P, Chen X (2007) Induction and preliminary characterization of a novel halophage SNJ1 from lysogenic Natrinema sp. F5. Can J Microbiol 53:1106–1110
Minakhin L, Goel M, Berdygulova Z, Ramanculov E, Florens L, Glazko G, Karamychev VN, Slesarev AI, Kozyavkin SA, Khromov I, Ackermann HW, Washburn M, Mushegian A, Severinov K (2008) Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23-45 and P74-26: siphoviruses with triplex-forming sequences and the longest known tails. J Mol Biol 378:468–480
Naryshkina T, Liu J, Florens L, Swanson SK, Pavlov AR, Pavlova NV, Inman R, Minakhin L, Kozyavkin SA, Washburn M, Mushegian A, Severinov K (2006) Thermus thermophilus Bacteriophage ϕYS40 genome and proteomic characterization of virions. J Mol Biol 364:667–677
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 13:1605–1612
Pietilä MK, Laurinavičius S, Sund J, Roine E, Bamford DH (2010) The single-stranded DNA genome of novel archaeal virus Halorubrum pleomorphic virus 1 is enclosed in the envelope decorated with glycoprotein spikes. J Virol 84:788–798
Porter K, Kukkaro P, Bamford JK, Bath C, Kivelä HM, Dyall-Smith ML, Bamford DH (2005) SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology 335:22–33
Porter K, Dyall-Smith ML (2008) Transfection of haloarchaea by the DNAs of spindle and round haloviruses and the use of transposon mutagenesis to identify non-essential regions. Mol Microbiol 70:1236–1245
Porter K, Russ BE, Yang J, Dyall-Smith ML (2008) The transcription programme of the protein-primed halovirus SH1. Microbiology 154:3599–3608
Porter K, Tang SL, Chen CP, Chiang PW, Hong MJ, Dyall-Smith M (2013) PH1: an archaeovirus of Haloarcula hispanica related to SH1 and HHIV-2. Archaea 2013:456318
Prangishvili D, Forterre P, Garrett RA (2006) Viruses of the archaea: a unifying view. Nat Rev Microbiol 4:837–848
Prangishvili D (2013) The wonderful world of archaeal viruses: a personal experience. Annu Rev Microbiol 67:565–585
Reiter WD, Palm P, Yeats S (1989) Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res 17:1907–1914
Rissanen I, Pawlowski A, Harlos K, Grimes JM, Stuart DI, Bamford JK (2012) Crystallization and preliminary crystallographic analysis of the major capsid proteins VP16 and VP17 of bacteriophage P23-77. Acta Crystallogr F 68:580–583
Rissanen I, Grimes JM, Pawlowski A, Mäntynen S, Harlos K, Bamford JK, Stuart DI (2013) Bacteriophage P23-77 capsid protein structures reveal the archetype of an ancient branch from a major virus lineage. Structure 21:718–726
Saren AM, Ravantti JJ, Benson SD, Burnett RM, Paulin L, Bamford DH, Bamford JK (2005) A snapshot of viral evolution from genome analysis of the Tectiviridae family. J Mol Biol 350:427–440
Shen P, Chen Y (1994) Plasmid from Halobacterium halobium and its restriction map. Yi Chuan Xue Bao 21:409–441
Strömsten NJ, Bamford DH, Bamford JK (2005) In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses. J Mol Biol 348:617–629
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M (2004) Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol 78:1954–1961
Xu L, Benson SD, Butcher SJ, Bamford DH, Burnett RM (2003) The receptor binding protein P2 of PRD1, a virus targeting antibiotic-resistant bacteria, has a novel fold suggesting multiple functions. Structure 11:309–322
Ye X, Ou J, Ni L, Shi W, Shen P (2003) Characterization of a novel plasmid from extremely halophilic Archaea: nucleotide sequence and function analysis. FEMS Microbiol Lett 221:53–57
Yu MX, Slater MR, Ackermann HW (2006) Isolation and characterization of Thermus bacteriophages. Arch Virol 151:663–679
Zhang Z, Liu Y, Wang S, Yang D, Cheng Y, Hu J, Chen J, Mei Y, Shen P, Bamford DH, Chen X (2012) Temperate membrane-containing halophilic archaeal virus SNJ1 has a circular dsDNA genome identical to that of plasmid pHH205. Virology 434:233–241
Acknowledgements
This work was supported by the Finnish Center of Excellence (CoE) program of the Acadamy of Finland 2012-2017 (CoE in Biological Interactions, #252411) and Acadamy of Finland personal grant to MJ (#251O13) and JKHB (#251106).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pawlowski, A., Rissanen, I., Bamford, J.K.H. et al. Gammasphaerolipovirus, a newly proposed bacteriophage genus, unifies viruses of halophilic archaea and thermophilic bacteria within the novel family Sphaerolipoviridae . Arch Virol 159, 1541–1554 (2014). https://doi.org/10.1007/s00705-013-1970-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-013-1970-6