Abstract
Chickpea chlorotic stunt virus (CpCSV), a proposed new member of the genus Polerovirus (family Luteoviridae), has been reported only from Ethiopia. In attempts to determine the geographical distribution and variability of CpCSV, a pair of degenerate primers derived from conserved domains of the luteovirus coat protein (CP) gene was used for RT-PCR analysis of various legume samples originating from five countries and containing unidentified luteoviruses. Sequencing of the amplicons provided evidence for the occurrence of CpCSV also in Egypt, Morocco, Sudan, and Syria. Phylogenetic analysis of the CP nucleotide sequences of 18 samples from the five countries revealed the existence of two geographic groups of CpCSV isolates differing in CP sequences by 8–10%. Group I included isolates from Ethiopia and Sudan, while group II comprised those from Egypt, Morocco and Syria. For distinguishing these two groups, a simple RFLP test using HindIII and/or PvuII for cleavage of CP-gene-derived PCR products was developed. In ELISA and immunoelectron microscopy, however, isolates from these two groups could not be distinguished with rabbit antisera raised against a group-I isolate from Ethiopia (CpCSV-Eth) and a group-II isolate from Syria (CpCSV-Sy). Since none of the ten monoclonal antibodies (MAbs) that had been produced earlier against CpCSV-Eth reacted with group-II isolates, further MAbs were produced. Of the seven MAbs raised against CpCSV-Sy, two reacted only with CpCSV-Sy and two others with both CpCSV-Sy and -Eth. This indicated that there are group I- and II-specific and common (species-specific) epitopes on the CpCSV CP and that the corresponding MAbs are suitable for specific detection and discrimination of CpCSV isolates. Moreover, CpCSV-Sy (group II) caused more severe stunting and yellowing in faba bean than CpCSV-Eth (group I). In conclusion, our data indicate the existence of a geographically associated variation in the molecular, serological and presumably biological properties of CpCSV.
Similar content being viewed by others
Introduction
Faba bean (Vicia faba L.), chickpea (Cicer arietinum L.), lentil (Lens culinaris Medik.), field pea (Pisum sativum L.) and grasspea (Lathyrus sativus L.), collectively called cool-season food legumes [26], constitute an important part of the human diet worldwide. They are a staple food for hundreds of millions of people, particularly in Asia and Africa. Virus diseases are among the major biotic constraints to the production of cool-season food legumes, which are most severely affected by yellowing and stunting diseases caused predominantly by luteoviruses [5]. Several viruses belonging to various taxa are known to be associated with yellowing and stunting diseases in countries of West Asia and North Africa (WANA). These include the nanovirus faba bean necrotic yellows virus (FBNYV), the mastrevirus chickpea chlorotic dwarf virus (CpCDV), and luteoviruses such as bean leaf roll virus (BLRV), soybean dwarf virus (SbDV) and viruses [e.g., turnip yellows virus (TuYV)] of the Beet western yellows virus (BWYV) subgroup [1, 9, 15–17, 19, 27].
Recently, a new member of the genus Polerovirus (family Luteoviridae) named chickpea chlorotic stunt virus (CpCSV) has been shown to naturally infect a range of cool-season food legumes in Ethiopia and to cause leaf yellowing and plant stunting in these legume crops [2]. The virus is transmitted by Aphis craccivora in the circulative (persistent) manner, and its experimental host range is apparently confined to a few cool-season food legume species. CpCSV shares a coat protein (CP) gene sequence identity of ~78% with groundnut rosette assistor virus (GRAV) which thus appears to be its closest relative [2].
Although there are several indications that luteoviruses prevail in cool-season food legumes in WANA countries, they often have not been accurately identified, chiefly due to the lack of specific antibodies and/or sequence information for serological and/or molecular detection, respectively. For the same reason, some authors have referred to the causal viruses only as a group [e.g., 1, 7, 8, 12, 18]. Even though first attempts at characterizing some of these unknown viruses provided molecular evidence for the existence of hitherto unrecognized luteoviruses in Morocco [9], Ethiopia and Sudan [3], the exact identity, biology and epidemiology of most of these luteoviruses that infect cool-season food legumes in the region are still enigmatic.
To obtain a better understanding of the geographic distribution and variability of CpCSV in WANA countries, the universal luteovirus primers described previously [2] were used for RT-PCR analysis of legume samples originating from various WANA countries and containing unknown luteoviruses. In this paper, we provide evidence for the occurrence of CpCSV in countries other than Ethiopia and for the existence of a geographically associated variation among CpCSV isolates differing not only in CP (molecular and serological) properties but presumably also in virulence.
Materials and methods
Origin of samples and isolates
From faba bean, chickpea, lentil, grasspea and fenugreek (Trigonella foenum-graecum) showing yellowing and stunting symptoms in the major legume production areas of Ethiopia in 2002, leaf samples were collected and preserved over CaCl2 (Table 1). Luteovirus-infected faba bean samples from Egypt and Morocco and chickpea samples from Sudan were selected from a collection of CaCl2-preserved samples that had formerly been analyzed by serological means at JKI, Braunschweig, Germany. The formerly used isolate CpCSV-FB from faba bean in Ambo, Ethiopia [2], referred to here as CpCSV-Eth (Et-fb-am in Table 1), was maintained in faba bean seedlings by vector transmission using Aphis craccivora Koch. Lyophilized tissue of faba bean leaves containing an unidentified luteovirus (SV-1-03) from Syria was kindly provided by S. G. Kumari and K. M. Makkouk, Aleppo, Syria. This virus, referred to here as CpCSV-Sy (Sy-fb1-03 in Table 1), was identified as a CpCSV isolate and reactivated by purifying virions from the dried tissue as described [2] and giving A. craccivora nymphs an acquisition access feeding for 48 h on purified virions using a membrane feeding system as described [15], followed by an inoculation access feeding on faba bean seedlings. Thereafter, CpCSV-Sy was maintained under glasshouse conditions using viruliferous A. craccivora nymphs for inoculating faba bean seedlings at 3- to 4-week intervals.
Serological analysis of samples
All samples were tested by triple antibody sandwich (TAS)-ELISA [10] using the various antibody combinations for broad-spectrum and specific detection of luteoviruses [2]. To find out whether the two groups of isolates differing in CP sequence also diverge in serological properties, CpCSV-Eth (group I) and -Sy (group II) were serologically analyzed in detail using antisera and MAbs raised ealier against CpCSV-Eth [2] and here against CpCSV-Sy in double antibody (DAS)-ELISA [6] and TAS-ELISA [10], respectively. ELISA reactions were considered positive when A405 nm values were at least two times higher than the mean A405 nm value for the non-infected controls. In addition, immunoelectron microscopy (IEM) decoration experiments were carried out as described [21] using purified virion preparations of, and the antisera to, both isolates. Differences in IEM decoration were only considered significant when the heterologous decoration titer differed from the homologous decoration titer by more than one dilution step in at least two separate experiments. Similarly, a range of luteovirus antisera available at the JKI collection, namely those to GRAV, BWYV, BLRV, SbDV, pea enation mosaic virus-1 (PEMV), potato leafroll virus (PLRV), cereal yellow dwarf virus (CYDV)-RPV, carrot red leaf virus (CtRLV), barley yellow dwarf virus-PAV (BYDV-PAV) and BYDV-MAV were used in the decoration experiments on purified preparations of the two isolates.
Virus purification and production of poly- and monoclonal antibodies
Purification of CpCSV-Sy virions and production of a rabbit antiserum to CpCSV-Sy was done using quantities and methods specified elsewhere [2]. IgG isolation, labeling of IgG with alkaline phosphatase (Roche, Germany), and DAS-ELISA were conducted as described [6]. For MAb production, each of two female Balb/c mice (2–3 months old) received two subcutaneous injections of 100 μl containing ~20 μg of CpCSV-Sy virions and 50% Freund’s incomplete adjuvant. Booster injections, fusion experiments, selection and cloning of hybridomas as well as isotyping of MAbs were conducted essentially as described [2, 4, 10]. Serial dilutions of culture supernatants ranging from 1:10 to 1:64,000 were tested in TAS-ELISA for determining MAb titers. The specificity of MAbs was assessed in Western blots using both purified CpCSV-Sy preparations and leaf extracts from CpCSV-Sy-infected and non-infected faba bean [29] as well as in (IEM) decoration experiments [21] using purified preparations of CpCSV-Sy virions.
Total RNA extraction, RT-PCR, cloning, and sequence analysis
A total of 30 luteovirus-positive samples (i.e., those reacting with MAb B-2-5G4 [14]) originating from five countries were used for amplification of CP sequences by reverse transcriptase (RT)-PCR. Further details of the samples from which CP gene sequences of CpCSV were amplified are presented in Table 1. Total RNA was extracted from CaCl2-preserved leaf samples using Nucleospin® Plant kit (Macherey-Nagel, Düren, Germany) following the manufacturer’s instructions, except that the desiccated leaf tissue was ground to a fine powder without liquid nitrogen using a pestle and mortar. A one-step RT-PCR was carried out essentially as described [2] using the degenerate primer pair (sense primer 5′-GCTCTAGAATTGTTAATGARTACGGTCG-3′ and antisense primer, 5′-CACGCGTCIACCTATTTIGGRTTITG-3′) derived from conserved terminal domains of the CP gene. Purification of PCR products, cloning and sequencing were done following standard procedures [25]. Sequence assembly, multiple alignment and pairwise identity analyses of nucleotide and amino acid sequences were carried out using DNAMAN (Lynnon Biosoft, Canada). Phylogenetic trees were constructed using the CLUSTAL_X program following alignment of sequences by neighbor-joining algorithms [28] and visualized using the TreeView program [24].
RFLP analysis
To develop an RFLP method for distinguishing the two groups of CpCSV isolates that differ in CP gene sequence, the PCR products obtained from the CP genes of the isolates with known CP sequences were digested using the restriction enzymes HindIII and PvuII (Fermentas). These were selected on the basis of analysis of unique restriction sites in the CP nucleotide sequences of each strain group of CpCSV. Restriction digestion was carried out separately for each enzyme in a reaction volume of 20 μl consisting of 5 μl PCR product, 2 μl of 10× restriction enzyme buffer (Fermentas), 0.3 μl of enzyme (10 U/μl) and 12.7 μl of H2O, followed by incubation at 37°C for 2 h using the specific buffers provided by the manufacturer.
Virulence of CpCSV-Eth and -Sy
Possible differences in symptom severity between the two groups of CpCSV isolates were studied by inoculating CpCSV-Eth [2] and -Sy separately onto faba bean, chickpea, lentil and field pea. Plants were inoculated by giving Aphis craccivora an acquisition access feeding of 2 days on infected faba bean plants, followed by an inoculation access feeding of 2 days on young seedlings of the four recipient plant species. Plants were kept in the glasshouse, inspected for symptom development at weekly intervals for up to 8 weeks and then analyzed for systemic infections using CpCSV-specific antibodies in DAS-ELISA. No attempts were made to establish correlations between symptom severity and virus titers in infected plants.
Results and discussion
The occurrence and molecular variability of CpCSV in legume samples from five WANA countries
Detection and identification of CpCSV in legume samples from Ethiopia provided some indications that it is a common virus in cool-season food legumes in this country [2; unpublished data]. This prompted us not only to study further CpCSV isolates from Ethiopia but also to determine to what extent CpCSV occurs in legume samples from other WANA countries. For this purpose we studied 30 legume samples that had given TAS-ELISA reactions with MAb B-2-5G4, a broad-spectrum MAb to luteoviruses [14], but had failed to react with virus-specific antibodies to BLRV, SbDV, or viruses of the BWYV subgroup (Table 2). This suggested that these ELISA-positive samples were infected either with CpCSV or other unrecognized luteoviruses.
To identify these unknown luteoviruses, the degenerate primers employed previously for CpCSV detection [2] were used for RT-PCR amplification of CP gene sequences from the samples. Out of the 30 ELISA-positive samples analyzed by RT-PCR, CP gene sequences of CpCSV were amplified from a total of 18 samples originating from Ethiopia, Egypt, Morocco, Sudan, and Syria (Table 2), indicating that CpCSV not only has a wide distribution in Ethiopia but also occurs in legume crops of other WANA countries. From the remaining 12 samples, either other luteovirus sequences were amplified [3] or no PCR product was obtained.
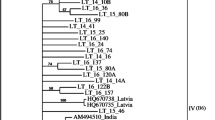
Although CpCSV sequences were obtained from 12 faba bean, three chickpea, one grasspea and two fenugreek samples (Table 1), there was no indication of host plant-specific variation in CP gene sequences when their sequences were compared (Fig. 1). However, analysis of the CP sequences from five WANA countries revealed a geographically associated variation among CpCSV isolates. The phylogenetic tree shown in Fig. 1 illustrates the relationships among the CpCSV isolates based on CP nucleotide (nt) sequences. Using the CP gene of GRAV, the closest relative of CpCSV [2], as an outgroup sequence, the tree reveals that ten isolates from Ethiopia and one isolate from Sudan form a distinct cluster (group I) that is clearly different from a second cluster (group II) consisting of one, five and one isolates from Egypt, Morocco and Syria, respectively. In pairwise comparisons of CP nt and amino acid (aa) sequences, the intergroup identities in nt and aa sequences ranged from 91 to 93% and from 89.4 to 92.1%, respectively. The CP gene identities between isolates within group I and within group II ranged from 95 to 100% and from 98 to 100%, respectively, for both nt and aa sequences.
Unrooted tree showing the phylogenetic relationship among the coat protein nucleotide sequences of 18 CpCSV isolates from five countries. The numbers refer to the serial number of each isolate as listed in Table 1. Groundnut rosette assistor virus (GRAV) sequence (acc. no. AF195828) was used as an outgroup. The sequences were aligned and neighbor-joining trees were constructed using the CLUSTAL_X program and viewed in TreeView. The number (in italics) at the major node is the bootstrap score for the division of the two groups of CpCSV isolates. The bar indicates the number of nt substitutions per site
On the basis of the available CP sequences, group-II isolates were thus not only less variable than group-I isolates but also less related (aa sequence identities of ~73%) to GRAV than group-I isolates (75.5–77.1%). This is illustrated in Fig. 2, which, on close examination of the CP sequences, shows that CpCSV-Eth (group I) and GRAV have nine identical amino acid positions that are absent from the CP of CpCSV-Sy (group II), whereas there is only one amino acid position unique for both CpCSV-Sy and GRAV CPs. Our observation that the intragroup variation of group-II isolates was lower than that of the group-I isolates is surprising, as the latter originated from Ethiopia (ten samples) and Sudan (one sample), two neighboring countries in northeast Africa, whereas group-II isolates were from Egypt and Syria (one sample each), two countries far away from Morocco, where the other five samples containing group-II isolates originated. Moreover, the samples containing group-I isolates were collected over a time period of 9 years (Table 1).
Alignment of the deduced coat protein amino acid sequence of a group-I and a group-II isolate of CpCSV from Ethiopia (CpCSV-Eth; acc. no. AY956384) and Syria (CpCSV-Sy; acc. no. EU541270), respectively, with that of groundnut rosette assistor virus (GRAV; acc. no. AF195828). Amino acid residues identical to, and different from, those of CpCSV-Eth are indicated by a dash and a letter, respectively. Single dots denote gaps. Identical amino acid residues specific for the pairs CpCSV-Eth/GRAV and CpCSV-Sy/GRAV are boxed using solid and dotted lines, respectively
Our data on CpCSV appear to be largely in line with the notion of a strict correlation between sequence variation and geographical origin of luteovirus isolates, as recently observed also for Sugarcane yellow leaf virus (ScYLV) [22] and Curcurbit aphid-borne yellows virus [30]. One explanation for this correlation might be the fact that luteoviruses are typically not seed-transmitted. The only exceptions to this rule might be luteoviruses in vegetatively propagated plants, for instance PLRV in potato [11, and references therein]. The close phylogenetic relationship between CpCSV and GRAV, their transmission by the same aphid species (A. craccivora), and the fact that they have an overlapping geographical distribution in Africa (e.g., in Ethiopia and Sudan) [2, 13, 23, this study] may indicate that the two viruses share a common ancestor in Africa, possibly in an unknown indigenous African legume species. It is possible that CpCSV isolates have coevolved with these legume plants and developed independently into different strains in geographical isolation.
RFLP test for discrimination of CpCSV isolates
In silico analysis of the CP sequences of the CpCSV group-I and -II isolates revealed unique restriction sites for each geographic group. RFLP analysis showed that all of the RT-PCR products obtained from seven group-I isolates studied were digested with PvuII, giving fragment sizes of about 430 and 170 bp, while those from the seven group-II isolates were not digested with PvuII (Fig. 3). Conversely, all amplicons from the group-II isolates were digested with HindIII, giving fragment sizes of 470 and 130 bp, whereas those from isolates of group I were not digested with this enzyme. Hence, either one or both of these two enzymes can be used for differentiating the two CpCSV groups on the basis of RFLP analyses of RT-PCR products.
Restriction fragment length polymorphisms of RT-PCR products from the coat protein gene of selected group-I and group-II isolates of CpCSV from five countries, digested with enzymes PvuII (a) and HindIII (b). Lane M shows the size markers, and the other lanes contain the digested PCR products from isolates from Ethiopia (1–6), Sudan (7), Morocco (8–12), Egypt (13) and Syria (14). The approximate sizes of the marker bands and the digested products are indicated at the left and right side of the figure, respectively
Serological properties of CpCSV isolates
The striking difference in CP aa sequences (8–10%) between the two geographic groups of CpCSV isolates studied here suggested that a serological differentiation of group-I from group-II isolates is possible. Since an antiserum and MAbs to a group-I isolate from Ethiopia (CpCSV-Eth) were available from a previous study [2], we purified the virions of CpCSV-Sy, a group-II isolate, and used them for polyclonal antibody (antiserum) and MAb production. This allowed us to test a representative isolate of each group in homologous and heterologous combinations of antigens and antibodies.
When the antiserum to CpCSV-Sy was used in DAS-ELISA, strong homologous reactions were observed using trapping IgG and enzyme-labeled antibodies at concentrations of 1 μg/ml and 1:1,000, respectively. Under these DAS-ELISA conditions, the CpCSV-Sy antiserum did not react with extracts from BLRV-, PEMV-, PLRV-, and SbDV-infected plants but reacted strongly with that of CpCSV-Eth and weakly with those from BMYV- and TuYV-infected plants (data not shown). Since serial dilutions of extracts from CpCSV-Eth- and -Sy-infected plants gave similar DAS-ELISA reactions with CpCSV-Eth and -Sy antisera in both homologous and heterologous combinations, these two isolates appeared serologically indistinguishable. Such a serological discrimination was also not possible in IEM as the homologous and heterologous decoration titers of the CpCSV-Eth and -Sy antisera with purified virions of each isolate were largely indistinguishable (data not shown). Similar to previous data for CpCSV-Eth [2], virions of CpCSV-Sy were decorated weakly with antibodies to CYDV-RPV, GRAV, PLRV and viruses of the BWYV subgroup, but no decoration was observed with antibodies against BLRV, SbDV, CtLRV, PEMV, BYDV-PAV and BYDV-MAV. Moreover, MAb 510H raised against an isolate of the BWYV subgroup gave an intermediate TAS-ELISA reaction with CpCSV-Sy (data not shown). In conclusion, no striking differences in the ELISA and IEM reactions of CpCSV-Eth and -Sy with the range of antisera tested were observed.
Monoclonal antibodies raised against CpCSV-Sy
Whereas CpCSV-Eth and -Sy appeared to be serologically indistinguishable in ELISA and IEM reactions with the range of antisera tested, CpCSV-Sy and the six other group-II isolates failed to react in TAS-ELISA with the ten MAbs to CpCSV-Eth, a group-I isolate. These MAbs had previously given strong TAS-ELISA reactions with all group-I isolates [2]. This observation suggested that all of the ten MAbs to CpCSV-Eth only recognize group I-specific epitopes. This prompted us to raise MAbs to CpCSV-Sy, a group-II isolate, in order to obtain MAbs that recognize not only group-II-specific eptopes but also epitopes shared by all CpCSV isolates.
From two fusion experiments, seven stable hybridoma lines secreting antibodies reacting with CpCSV-Sy but differing in luteovirus specificity were obtained. Three, one, and three of the MAbs were typed as IgG2a, IgG2b, and IgG1, respectively (Table 3), confirming the monoclonality of the antibodies. The MAbs gave weak to strong reactions with CpCSV-Sy (Table 3). However, when these MAbs were tested against CpCSV-Eth and the three other luteoviruses BLRV, PLRV and TuYV (a virus of the BWYV subgroup), MAbs 5-5B8 and 5-6G8 reacted only with CpCSV-Sy. In contrast, MAbs 5-1F10, 5-2B8, 5-3D5, and 5-6F11 reacted with both CpCSV-Sy and -Eth, but two of them (MAbs 5-1F10 and 5-6F11) gave a weak reaction also with TuYV. There was only one MAb (6-4E10) reacting with CpCSV-Sy and TuYV. None of the MAbs reacted with BLRV and PLRV.
To confirm the specificity of the MAbs, they were used for IEM decoration and Western blot experiments (Table 3). However, a visible decoration reaction with purified CpCSV-Sy virions was not readily produced with any of the MAbs. When immunogold labeling was used for revealing weak decoration reactions of MAbs with CpCSV-Sy virions, only MAbs 5-2B8 and 5-5B8 were shown to bind to virions. Immunogold labeling of the CpCSV-Sy particles with the other five MAbs produced either very low numbers of gold-labeled particles (only MAb 5-6F11) or none at all (four MAbs). In Western blot analysis of purified CpCSV-Sy virions and extracts from CpCSV-infected and non-inoculated plants, only MAb 5-6G8 gave a Western blot reaction similar in strength to the polyclonal antibodies, while the other six MAbs failed to react (results not shown).
There were striking differences in the titers of the MAb-containing culture supernatants (Table 3). For routine use in TAS-ELISA, culture supernatant dilutions of 1:50 to 1:4,000 were found to give the best signal-to-noise ratio. Use of polyclonal IgG to CpCSV-Sy or -Eth as capture antibody and each of the strongly (+++) reacting MAbs as detecting antibodies permitted more sensitive detection of CpCSV antigen in TAS-ELISA than the use of the polyclonal antibodies alone in DAS-ELISA (data not shown). Although the MAbs raised here against CpCSV-Sy have not been tested yet against a range of group-I and -II isolates of CpCSV, the available data suggest that MAbs 5-2B8 and 5-3D8 are suitable for specific TAS-ELISA detection of any CpCSV isolate. In contrast, MAb 5-5B8 and the previously produced MAbs to CpCSV-Eth [2] seem capable of specific TAS-ELISA detection of group-II and -I isolates, respectively, permitting discrimination and typing of CpCSV isolates.
Biological properties of group-I and -II isolates
By using a membrane feeding system for allowing Aphis craccivora nymphs an acquisition access to purified virions of CpCSV-Sy, we were able not only to establish a virus culture of this isolate but also to demonstrate infectivity of CpCSV particles. In this way, we substantiated the role of CpCSV in the etiology of the yellowing and stunting diseases of cool-season food legumes and fulfilled Koch’s postulates.
In subsequent inoculation experiments, Aphis craccivora was used for vector inoculation of four legume species (C. arietinum, L. culinaris, P. sativum and V. faba) with both CpCSV-Eth and -Sy. Under glasshouse conditions, they induced indistinguishable symptoms in three legume species, as infections in field pea were latent and the severities of leaf yellowing and plant stunting induced by each isolate in chickpea and lentil were similar. In faba bean, however, the symptomatology of the two isolates was strikingly distinct in three separate experiments (Fig. 4a, b). CpCSV-Eth caused a slight growth reduction and a mild leaf yellowing that was most obvious only in older leaves. In contrast, faba bean plants infected with CpCSV-Sy were severely stunted and developed leaves showing a conspicuous yellowing and a leathery texture in both old and young leaves, often followed by plant death.
Variation in plant stunting (a) and leaf yellowing (b) caused by the Syrian isolate Sy-fb1-03 (CpCSV-Sy; left) and the Ethiopian isolate Et-fb-am (CpCSV-Eth; middle) in the faba bean cultivar Condor under glasshouse conditions. An uninoculated control plant and leaf are shown on the right. Photographs were taken ~6 weeks after inoculation
Due to the inavailability of further CpCSV isolates for biological studies, however, we were unable to substantiate the virulence difference between the two groups of CpCSV isolates. Similarly, it is currently not known whether the two groups of CpCSV isolates also differ in aphid vector specificity. CpCSV-Sy, a group-II isolate, was transmitted by A. craccivora as efficiently as the group-I isolate CpCSV-Eth (data not shown), which was not vectored by Acyrthosiphon pisum, Aphis fabae or Myzus persicae in a previous study [2]. Here, no attempts were made to use these three aphid species for vector transmission of CpCSV-Sy.
Concluding remarks
After our previous report on CpCSV infections of faba bean, chickpea, grasspea and fenugreek in Ethiopia [2], we here provide the first evidence for the occurrence of CpCSV in faba bean and chickpea in Sudan, Egypt, Morocco and Syria and the existence of two distinct groups of CpCSV isolates that appear to be geographically separated. In addition to these molecular data, the differential reactions of the available MAbs with the two groups of CpCSV isolates also provide evidence for differences in serological properties between the two groups of CpCSV isolates. The clear difference in symptom severity observed in faba bean for a representative isolate of each group may be a first indication that the two geographically distinct strain groups of CpCSV differ not only genetically and serologically but also biologically (in virulence).
Despite the limited number of samples used for sequence analysis, CpCSV sequences were amplified from the majority (18/30) of the luteovirus-positive samples analyzed (Table 2). This suggests that the virus is widespread in cool-season food legumes grown in WANA countries, probably being more prevalent than the other luteoviruses known so far from the region, such as BLRV and SbDV [1, 7, 18, 19, 27]. The observed variability within the sequences of the two groups, together with its detection in samples collected as early as 1994 from Egypt and Morocco (Table 1), indicates that the virus has not emerged only very recently. It has probably been infecting legume plants for decades or centuries but has remained undetected and/or has been incorrectly identified as one of the other legume luteoviruses, possibly due to the lack of appropriate diagnostic tools or the use of cross-reacting antibodies. Further studies on both the incidence and variability of CpCSV in several WANA countries or adjacent regions are required for determining its economic importance.
Although results of serological surveys have contributed to a growing awareness of the importance of luteoviruses in legume crops in WANA countries, there have also been indications that the use of serological methods is not sufficiently reliable for the identification of luteovirus species due to the fact that different luteoviruses share a number of immunodominant epitopes [e.g., 2, 9, 20]. On the other hand, there is very little sequence information on legume luteoviruses from the WANA region in the database. The sequence information generated here and in previous work [2, 3] forms the basis for further research on CpCSV and other luteoviruses, including design of virus- or strain-specific primers. In addition, the degenerate primer pair used in this study for the amplification of CP gene sequences was useful in our hands for detection of not only CpCSV but also other luteo- and poleroviruses [3]. Therefore, they appear suitable for obtaining CP gene sequences of further unknown luteo- and poleroviruses. Finally, their use in combination with RFLP analysis of RT-PCR products and/or virus- or strain-specific MAbs produced here and previously [2] will certainly facilitate future studies on the identification, ecology and geographical distribution of CpCSV and its strains throughout the world.
The information presented here on the existence and geographically associated variation of CpCSV strains will help breeders in their attempts to incorporate virus resistance to the prevalent CpCSV strains in their region or country, since sources of resistance selected in one country may not necessarily be suitable for other countries where different virus strains can occur.
References
Abraham AD, Makkouk KM, Gorfu D, Lencho AG, Ali K, Tadesse N, Yusuf A, Lencho A (2000) Survey of faba bean (Vicia faba L.) virus diseases in Ethiopia. Phytopathol Mediterr 39:277–282
Abraham AD, Menzel W, Lesemann D-E, Varrelmann M, Vetten HJ (2006) Chickpea chlorotic stunt virus: a new polerovirus infecting cool-season food legumes in Ethiopia. Phytopathology 96:437–446
Abraham AD, Varrelmann M, Vetten HJ (2008) Molecular evidence for the occurrence of two new luteoviruses in cool season food legumes in Northeast Africa. Afr J Biotechnol 7:414–420
Adam G, Lesemann D-E, Vetten HJ (1991) Monoclonal antibodies against tomato spotted wilt virus: characterization and application. Ann Appl Biol 118:87–104
Bos L, Hampton RO, Makkouk KM (1988) Viruses and virus diseases of pea, lentil, faba bean and chickpea. In: Summerfield RJ (ed) World crops; cool season food legumes. Kluwer, Dordrecht, pp 591–615
Clark MF, Adams AN (1977) Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J Gen Virol 34:475–483
Fortass MF, Bos L (1991) Survey of faba bean (Vicia faba L) for viruses in Morocco. Neth J Plant Pathol 97:369–380
Fortass MF, van der Wilk F, Goldbach RW, Bos L, van den Heuvel JFJM (1996) Diversity of viruses infecting faba bean in Morocco and their detection by the polymerase chain reaction. Agronomie 16:61–68
Fortass MF, van der Wilk F, van den Heuvel JFJM, Goldbach RW (1997) Molecular evidence for the occurrence of beet western yellows virus on chickpea in Morocco. Eur J Plant Pathol 103:481–484
Franz A, Makkouk KM, Katul L, Vetten HJ (1996) Monoclonal antibodies for the detection and differentiation of faba bean necrotic yellows virus isolates. Ann Appl Biol 128:255–268
Guyader S, Ducray DG (2002) Sequence analysis of Potato leaf roll virus isolates reveals genetic stability, major evolutionary events and differential selection pressure between overlapping reading frame products. J Gen Virol 83:1799–1807
Horn NM, Makkouk KM, Kumari SG, van den Heuvel JFJM, Reddy DVR (1995) Survey of chickpea (Cicer arietinum L.) for chickpea stunt disease and associated viruses in Syria, Turkey and Lebanon. Phytopathol Mediterr 34:192–198
Hull R, Adams AN (1968) Groundnut rosette and its assistor virus. Ann Appl Biol 62:139–145
Katul L (1992) Serologische und molekularbiologische Characterisierung des bean leaf roll virus (BLRV) und des faba bean necrotic yellows virus (FBNYV). PhD dissertation, University of Göttingen, Germany, p 115
Katul L, Vetten HJ, Maiss E, Makkouk KM, Lesemann D-E, Casper R (1993) Characterization and serology of virus-like particles associated with faba bean necrotic yellows. Ann Appl Biol 123:629–647
Makkouk KM, Dafalla G, Hussein M, Kumari SG (1995) The natural occurrence of chickpea chlorotic dwarf geminivirus in chickpea and faba bean in the Sudan. J Phytopathol 143:465–466
Makkouk KM, Damsteegt V, Johnstone GR, Katul L, Lesemann D-E, Kumari SG (1997) Identification and some properties of soybean dwarf luteovirus affecting lentil in Syria. Phytopathol Mediterr 36:135–144
Makkouk KM, Kumari SG, Shahraeen N, Fazlali Y, Farzadfar S, Ghotbi T, Mansouri RA (2003) Identification and seasonal variation of viral diseases of chickpea and lentil in Iran. J Plant Dis Prot 110:157–169
Makkouk KM, Rizkallah L, Madkour M, El-Sherbeny M, Kumari SG, Amriti AW, Solh MB (1994) Survey of faba bean (Vicia faba L) for viruses in Egypt. Phytopathol Mediterr 33:207–211
Martin RR, D’Arcy CJ (1990) Relationship among luteoviruses based on nucleic acid hybridisation and serological studies. Intervirology 31:23–30
Milne RG, Lesemann D-E (1984) Immunosorbent electron microscopy in plant virus studies. Methods in Virology VIII:85–101
Moonan F, Mirkov TE (2002) Analysis of genomic diversity among North, South and Central American isolates of Sugarcane yellow leaf virus: evidence for Colombian origins and for intraspecific spatial phylogenetic variation. J Virol 76:1339–1348
Naidu RA, Kimmins FM, Deom CM, Subrahmanyam P, Chiyembekeza AJ, van der Merwe PJA (1999) Groundnut rosette: a virus disease affecting groundnut production in sub-Saharan Africa. Plant Dis 83:700–709
Page GD (1996) Treeview: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Second edition, Plainview. Cold Spring Harbour Laboratory Press, New York
Summerfield RJ (ed) (1988) World crops: cool season food legumes. Kluwer, The Netherlands, p 1179
Tadesse N, Ali K, Gorfu D, Yusuf A, Abraham A, Ayalew M, Lencho A, Makkouk KM, Kumari SG (1999) Survey of chickpea and lentil virus diseases in Ethiopia. Phytopathol Mediterr 38:149–158
Thompson JD, Gibson TB, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucl Acids Res 25:4876–4882
Vetten HJ, Lesemann D-E, Maiss E (1992) Serotype A and B strains of bean common mosaic virus are two distinct potyviruses. Arch Virol, Suppl 5:415–431
Xiang HY, Shang QX, Han CG, Li DW, Yu JL (2008) Complete sequence analysis reveals two distinct poleroviruses infecting cucurbits in China. Arch Virol 153:1155–1160
Acknowledgments
This work was supported partly by a scholarship from German Academic Exchange Service (DAAD) to Adane D. Abraham. The excellent technical assistance of Christina Maass and Angelika Sieg-Müller is greatly appreciated. We thank Drs. K. M. Makkouk and S. G. Kumari for the faba bean sample containing the isolate Sy-fb1-03 from Syria. We gratefully acknowledge the gifts of antisera and antibodies from all those colleagues listed in Table 1 of [2].
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Abraham, A.D., Menzel, W., Varrelmann, M. et al. Molecular, serological and biological variation among chickpea chlorotic stunt virus isolates from five countries of North Africa and West Asia. Arch Virol 154, 791–799 (2009). https://doi.org/10.1007/s00705-009-0374-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-009-0374-0