Abstract
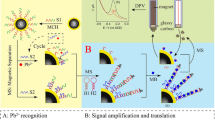
A highly sensitive and selective fluorescence method has been conducted for the detection of Hg2+ based on aminophenylboronic acid–modified carboxyl magnetic beads (CMB@APBA) and CRISPR/Cas12a system mediated by glyoxal caged nucleic acid (gcDNA). As a bi-functional DNA linker, gcDNA offers advantages of simultaneous recognition by boronic acid and complementary DNA/RNA. Under acidic condition, gcDNA can be immobilized on CMB@APBA through the formation of borate ester bond. The formed boric acid–esterified gcDNA can further bind with complementary CRISPR RNA through A-T base pairing to activate Cas12a with kcat/Km ratio of 3.4 × 107 s−1 M−1, allowing for amplified signal. Hg2+ can specifically combine with CMB@APBA, resulting in the release of gcDNA from CMB@APBA and the following inhibition on the activation of CRISPR/Cas12a system around magnetic bead. Under optimal conditions, the method exhibits a linear range from 20 to 250 nM, with a detection limit of 2.72 nM. The proposed method can detect Hg2+ in milk and tea beverages, providing a great significance for on-site monitoring of Hg2+ contamination in food.
Graphical abstract








Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
Guo Y, Werbel T, Wan S et al (2016) Potent antigen-specific immune response induced by infusion of spleen cells coupled with succinimidyl-4-(N-maleimidomethyl cyclohexane)-1-carboxylate (SMCC) conjugated antigens. Int Immunopharmacol 31:158–168. https://doi.org/10.1016/j.intimp.2015.12.023
Qing M, Xie S, Cai W et al (2018) Click chemistry reaction-triggered 3D DNA walking machine for sensitive electrochemical detection of copper ion. Anal Chem 90(19):11439–11445. https://doi.org/10.1021/acs.analchem.8b02555
Cao Y, Yu X, Han B et al (2021) In situ programmable DNA circuit-promoted electrochemical characterization of stemlike phenotype in breast cancer. J Am Chem Soc 143(39):16078–16086. https://doi.org/10.1021/jacs.1c06436
Knutson SD, Sanford AA, Swenson CS et al (2020) Thermoreversible control of nucleic acid structure and function with glyoxal caging. J Am Chem Soc 142(41):17766–17781. https://doi.org/10.1021/jacs.0c08996
Chen HJC, Chang YL, Teng YC et al (2017) A stable isotope dilution nanoflow liquid chromatography tandem mass spectrometry assay for the simultaneously detection and quantification of glyoxal-induced DNA cross-linked adducts in leukocytes from diabetic patients. Anal Chem 89(24):13082–13088. https://doi.org/10.1021/acs.analchem.6b04296
Otsuka H, Uchimura E, Koshino H et al (2004) Anomalous binding profile of phenylboronic acid with N-acetylneuraminic acid (Neu5Ac) in aqueous solution with varying pH. J Am Chem Soc 125(12):3493–3502
Zhang J, Liu Y, Wang X et al (2015) Electrochemical assay of α-glucosidase activity and the inhibitor screening in cell medium. Biosens Bioelectron 74:666–672. https://doi.org/10.1016/j.bios.2015.07.023
Zhang J, Lv J, Wang X et al (2015) Integration of chemoselective ligation with enzymespecific catalysis: saccharic colorimetric analysis using aminooxy/hydrazine-functionalized gold nanoparticles. Nano Res 8:3853–3863. https://doi.org/10.1007/s12274-015-0885-9
Zhang J, Liu Y, Lv J, Li G (2015) A colorimetric method for α-glucosidase activity assay and its inhibitor screening based on aggregation of gold nanoparticles induced by specific recognition between phenylenediboronic acid and 4-aminophenyl-α-d-glucopyranoside. Nano Res 8:920–930. https://doi.org/10.1007/s12274-014-0573-1
Kaback DB, Angerer LM, Davidson V (1980) Improved methods for the formation and stabilization of R-loops. Nucleic Acids Res 12(21):8235–8251
Cheng L, Yang F, Tang L et al (2022) Electrochemical evaluation of tumor development via cellular interface supported CRISPR/Cas trans-cleavage. Research. https://doi.org/10.34133/2022/9826484
Zhang J, Sheng A, Wang P et al (2021) Mxene coupled with crispr-cas12a for analysis of endotoxin and bacteria. Anal Chem 93(10):4676–4681. https://doi.org/10.1021/acs.analchem.1c00371
Yu Y, Li W, Gu X et al (2022) Inhibition of CRISPR-Cas12a trans-cleavage by lead (II)-induced G-quadruplex and its analytical application. Food Chem 378:131802. https://doi.org/10.1016/j.foodchem.2021.131802
Zhu Y, Zhu J, Gao Y et al (2023) Electrochemical determination of flap endonuclease 1 activity amplified by CRISPR/Cas12a trans-cleavage**. ChemElectroChem 10(8):e202300020. https://doi.org/10.1002/celc.202300020
Chen JS, Ma E, Harrington LB et al (2018) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360(6387):436–439. https://doi.org/10.1126/science.aar6245
Stella S, Mesa P, Thomsen J et al (2018) Conformational activation promotes CRISPR-Cas12a catalysis and resetting of the endonuclease activity. Cell 175(7):1856–1871. https://doi.org/10.1016/j.cell.2018.10.045
Li SY, Cheng QX, Liu JK et al (2018) CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res 28(4):491–493. https://doi.org/10.1038/s41422-018-0022-x
Dehghani Z, Nguyen T, Golabi M et al (2021) Magnetic beads modified with Pt/Pd nanoparticle and aptamer as a catalytic nano-bioprobe in combination with loop mediated isothermal amplification for the on-site detection of Salmonella typhimurium in food and fecal samples. Food Control 121:107664. https://doi.org/10.1016/j.foodcont.2020.107664
Yang S, Ouyang H, Su X et al (2016) Dual-recognition detection of Staphylococcus aureus using vancomycin-functionalized magnetic beads as concentration carriers. Biosens Bioelectron 78:174–180. https://doi.org/10.1016/j.bios.2015.11.041
Deng M, Li W, Chen Y et al (2022) Detection of fumonisin B1 by aptamer-functionalized magnetic beads and ultra-performance liquid chromatography. Microchem J 178:107346. https://doi.org/10.1016/j.microc.2022.107346
Li S, Qin Y, Zhong G et al (2018) Highly efficient separation of glycoprotein by dual-functional magnetic metal-organic framework with hydrophilicity and boronic acid affinity. ACS Appl Mater Interfaces 10(33):27612–27620. https://doi.org/10.1021/acsami.8b07671
Wang Z, Liu J, Chen G et al (2022) An integrated system using phenylboronic acid functionalized magnetic beads and colorimetric detection for Staphylococcus aureus. Food Control 133:108633. https://doi.org/10.1016/j.foodcont.2021.108633
Feng S, Zhang A, Wu F et al (2022) In-situ growth of boronic acid-decorated metal-organic framework on Fe3O4 nanospheres for specific enrichment of cis-diol containing nucleosides. Anal Chim Acta 1206:339772. https://doi.org/10.1016/j.aca.2022.339772
Zou WS, Ye CH, Wang YQ et al (2018) A hybrid ratiometric probe for glucose detection based on synchronous responses to fluorescence quenching and resonance light scattering enhancement of boronic acid functionalized carbon dots. Sens Actuators B Chem 271:54–63. https://doi.org/10.1016/j.snb.2018.05.115
Chatterjee A, Banerjee M, Khandare DG et al (2017) Aggregation-induced emission-based chemodosimeter approach for selective sensing and imaging of Hg(II) and methylmercury species. Anal Chem 89(23):12698–12704. https://doi.org/10.1021/acs.analchem.7b02663
Wang H, Wang X, Liang M et al (2020) A boric acid-functionalized lanthanide metal-organic framework as a fluorescence “turn-on” probe for selective monitoring of Hg2+ and CH3Hg+. Anal Chem 92(4):3366–3372. https://doi.org/10.1021/acs.analchem.9b05410
Hajri AK, Jamoussi B, Albalawi AE et al (2022) Designing of modified ion-imprinted chitosan particles for selective removal of mercury (II) ions. Carbohydr Polym 286:119207. https://doi.org/10.1016/j.carbpol.2022.119207
Yang J, Zhang Y, Guo J et al (2020) Nearly monodisperse copper selenide nanoparticles for recognition, enrichment, and sensing of mercury ions. ACS Appl Mater Interfaces 12(35):39118–39126. https://doi.org/10.1021/acsami.0c09865
Guo X, Huang J, Wei Y et al (2020) Fast and selective detection of mercury ions in environmental water by paper-based fluorescent sensor using boronic acid functionalized MoS2 quantum dots. J Hazard Mater 381:120969. https://doi.org/10.1016/j.jhazmat.2019.120969
Behbahani M, Bide Y, Salarian M et al (2014) The use of tetragonal star-like polyaniline nanostructures for efficient solid phase extraction and trace detection of Pb(II) and Cu(II) in agricultural products, sea foods, and water samples. Food Chem 158:14–19. https://doi.org/10.1016/j.foodchem.2014.02.110
Funding
This work is sponsored by the Special National Key Research and Development Plan (Grant No. 2018YFC160440), the Science and Technology Commission of Shanghai Municipality (Grant No. 20392001800), and the National Natural Science Foundation of China (Grant No. 31671923).
Author information
Authors and Affiliations
Contributions
Juan Zhang: conceived the project; Ying Yu: designed and coordinated the experiments; Ying Yu, Yuan Zhang, and Lelin Qian: performed experiments; Ying Yu, Wenhui Li, and Qin Mi: analyzed results; Ying Yu and Juan Zhang: wrote the manuscript; and Juan Zhang and Zhengwu Wang: funding acquisition. All authors reviewed and commented on the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, Y., Zhang, Y., Chen, X. et al. Bi-functionality of glyoxal caged nucleic acid coupled with CRISPR/Cas12a system for Hg2+ determination . Microchim Acta 191, 120 (2024). https://doi.org/10.1007/s00604-024-06196-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-024-06196-5