Abstract
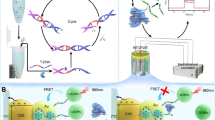
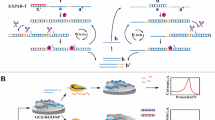
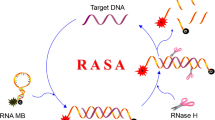
A novel electrochemical biosensor that combines the CRISPR-Cas12a system with a gold electrode is reported for the rapid and sensitive detection of microphthalmia-associated transcription factor (MITF). The biosensor consists of a gold electrode modified with DNA1, which contains the target sequence of MITF and is labeled with ferrocene, an electroactive molecule. The biosensor also includes hairpin DNA, which has a binding site for MITF and can hybridize with helper DNA to form a double-stranded complex that activates CRISPR-Cas12a. When MITF is present, it binds to hairpin DNA and prevents its hybridization with helper DNA, thus inhibiting CRISPR-Cas12a activity and preserving the DPV signal of ferrocene. When MITF is absent, hairpin DNA hybridizes with helper DNA and activates CRISPR-Cas12a, which cleaves DNA1 and releases ferrocene, thus reducing the DPV signal. The biosensor can detect MITF with high sensitivity (with an LOD of 8.14 fM), specificity, and accuracy in various samples, such as cell nuclear extracts and human serum. The biosensor can also diagnose and monitor melanocyte-related diseases and melanin production. This work provides a simple, fast, sensitive, and cost-effective biosensor for MITF detection and a valuable tool for applications in genetic testing, disease diagnosis, and drug screening.
Graphical abstract






Similar content being viewed by others
Data availability
The data and materials used in this research are available from the corresponding author upon reasonable request.
Change history
12 February 2024
A Correction to this paper has been published: https://doi.org/10.1007/s00604-024-06190-x
References
Gaggioli C, Buscà R, Abbe P, Ortonne J-P, Ballotti R (2003) Microphthalmia-associated transcription factor (MITF) is required but is not sufficient to induce the expression of melanogenic genes. Pigment Cell Res 16(4):374–382. https://doi.org/10.1034/j.1600-0749.2003.00057.x
Koyanagi K, O’Day SJ, Gonzalez R, Lewis K, Robinson WA, Amatruda TT et al (2006) Microphthalmia transcription factor as a molecular marker for circulating tumor cell detection in blood of melanoma patients. Clin Cancer Res 12(4):1137–1143. https://doi.org/10.1158/1078-0432.Ccr-05-1847
Tanwar J, Sharma A, Saurav S, Shyamveer, Jatana N, Motiani RK (2022) MITF is a novel transcriptional regulator of the calcium sensor STIM1: significance in physiological melanogenesis. J Biol Chem 298(12). https://doi.org/10.1016/j.jbc.2022.102681
Lee J, Hyeon DY, Hwang D (2020) Single-cell multiomics: technologies and data analysis methods. Exp Mol Med 52(9):1428–1442. https://doi.org/10.1038/s12276-020-0420-2
Wang R, He Y, Robinson V, Yang Z, Hessler P, Lasko LM et al (2018) Targeting lineage-specific MITF pathway in human melanoma cell lines by A-485, the selective small-molecule inhibitor of p300/CBP. Mol Cancer Ther 17(12):2543–2550. https://doi.org/10.1158/1535-7163.Mct-18-0511
Zhang K, Fan Z, Yao B, Zhang T, Ding Y, Zhu S et al (2021) Entropy-driven electrochemiluminescence ultra-sensitive detection strategy of NF-κB p50 as the regulator of cytokine storm. Biosens Bioelectron 176:112942. https://doi.org/10.1016/j.bios.2020.112942
Ding Y, Fan Z, Yao B, Xu D, Xie M, Zhang K (2021) Nanoparticle-based fluorescence probe for detection of NF-κB transcription factor in single cell via steric hindrance. Microchim Acta 188(7):226. https://doi.org/10.1007/s00604-021-04878-y
Zhao L, Li T, Xu X, Xu Y, Li D, Song W et al (2023) Polyhedral Au nanoparticle/MoOx heterojunction-enhanced ultrasensitive dual-mode biosensor for miRNA detection combined with a nonenzymatic cascade DNA amplification circuit. Anal Chem 95(24):9271–9. https://doi.org/10.1021/acs.analchem.3c01062
Zhou H, Zhang J, Li B, Liu J, Xu J-J, Chen H-Y (2021) Dual-mode SERS and electrochemical detection of miRNA based on popcorn-like gold nanofilms and toehold-mediated strand displacement amplification reaction. Anal Chem 93(15):6120–7. https://doi.org/10.1021/acs.analchem.0c05221
Wen X, Zhuo C, Wei J, Gong Y, Tang Q, Liao X et al (2023) Construction of a fluorescence sensing platform based on RISC and CRISPR/Cas12a for the assay of Ago2 enzyme activity. Microchem J 194:109284. https://doi.org/10.1016/j.microc.2023.109284
Yang Z, Guo Z, Yuan H, Li Y, Hu Y, Li X-Q et al (2023) Ultrasensitive detection of methicillin-resistant Staphylococcus aureus using a T7 exonuclease-assisted PAM-free dual CRISPR-Cas12a biosensor. Sens Actuators B Chem 396:134568. https://doi.org/10.1016/j.snb.2023.134568
Qing M, Sun Z, Wang L, Du SZ, Zhou J, Tang Q et al (2021) CRISPR/Cas12a-regulated homogeneous electrochemical aptasensor for amplified detection of protein. Sens Actuators B Chem 348:130713. https://doi.org/10.1016/j.snb.2021.130713
Yuan G, Xia X, Zhang J, Huang J, Xie F, Li X et al (2023) A novel “signal on-off-super on” sandwich-type aptamer sensor of CRISPR-Cas12a coupled voltage enrichment assay for VEGF detection. Biosens Bioelectron 221:114424. https://doi.org/10.1016/j.bios.2022.114424
Qiu M, Zhou X-M, Liu L (2022) Improved strategies for CRISPR-Cas12-based nucleic acids detection. J Anal Test 6(1):44–52. https://doi.org/10.1007/s41664-022-00212-4
Yang F, Wang W, Zhang M, Tao W, Wang Y, Shi J et al (2022) CRISPR/Cas12a-mediated electrochemiluminescence platform for environmental and human serum SARS-CoV-2 RNA monitoring using a self-enhanced ruthenium complex linked to zeolitic imidazole framework-8. Environ Sci Nano 9(9):3417–3426. https://doi.org/10.1039/d2en00595f
Qing M, Chen SL, Sun Z, Fan Y, Luo HQ, Li NB (2021) Universal and programmable rolling circle amplification-CRISPR/Cas12a-mediated immobilization-free electrochemical biosensor. Anal Chem 93(20):7499–7507. https://doi.org/10.1021/acs.analchem.1c00805
Zhu Z, Guo Y, Wang C, Yang Z, Li R, Zeng Z et al (2023) An ultra-sensitive one-pot RNA-templated DNA ligation rolling circle amplification-assisted CRISPR/Cas12a detector assay for rapid detection of SARS-CoV-2. Biosens Bioelectron 228:115179. https://doi.org/10.1016/j.bios.2023.115179
Li S-Y, Cheng Q-X, Wang J-M, Li X-Y, Zhang Z-L, Gao S et al (2018) CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov 4(1):20. https://doi.org/10.1038/s41421-018-0028-z
Zhang K, Fan Z, Ding Y, Zhu S, Xie M, Hao N (2022) Exploring the entropy-driven amplification reaction and trans-cleavage activity of CRISPR-Cas12a for the development of an electrochemiluminescence biosensor for the detection of the SARS-CoV-2 RdRp gene in real samples and environmental surveillance. Environ Sci Nano 9(1):162–172. https://doi.org/10.1039/d1en00645b
Oesinghaus L, Simmel FC (2021) Controlling Gene expression in mammalian cells using multiplexed conditional guide RNAs for Cas12a**. Angew Chem Int Ed 60(44):23894–23902. https://doi.org/10.1002/anie.202107258
Xiong Q, Xie C, Zhang Z, Liu L, Powell JT, Shen Q et al (2020) DNA origami post-processing by CRISPR-Cas12a. Angew Chem Int Ed 59(10):3956–3960. https://doi.org/10.1002/anie.201915555
Zhang M, Wang H, Wang H, Wang F, Li Z (2021) CRISPR/Cas12a-assisted ligation-initiated loop-mediated isothermal amplification (CAL-LAMP) for highly specific detection of microRNAs. Anal Chem 93(22):7942–7948. https://doi.org/10.1021/acs.analchem.1c00686
Zhang K, Fan Z, Ding Y, Xie M (2022) A pH-engineering regenerative DNA tetrahedron ECL biosensor for the assay of SARS-CoV-2 RdRp gene based on CRISPR/Cas12a trans-activity. Chem Eng J 429:132472. https://doi.org/10.1016/j.cej.2021.132472
Fan Z, Ding Y, Yao B, Wang J, Zhang K (2021) Electrochemiluminescence platform for transcription factor diagnosis by using CRISPR–Cas12a trans-cleavage activity. Chem Commun 57(65):8015–8018. https://doi.org/10.1039/D1CC03071J
Wang X-Y, Li Y, Lv S, Bi S (2022) pH-responsive magnetic I-motif container coupled with DNA walker for construction of dual-signal electrochemical biosensor. J Anal Test 6(1):12–19. https://doi.org/10.1007/s41664-021-00205-9
Adornetto G, Porchetta A, Palleschi G, Plaxco KW, Ricci F (2015) A general approach to the design of allosteric, transcription factor-regulated DNAzymes. Chem Sci 6(7):3692–3696. https://doi.org/10.1039/C5SC00228A
Liu J-J, Song X-R, Wang Y-W, Chen G-N, Yang H-H (2012) A graphene oxide (GO)-based molecular beacon for DNA-binding transcription factor detection. Nanoscale 4(12):3655–3659. https://doi.org/10.1039/C2NR30499F
Zhu D, Wang L, Xu X, Jiang W (2016) Label-free and enzyme-free detection of transcription factors with graphene oxide fluorescence switch-based multifunctional G-quadruplex-hairpin probe. Biosens Bioelectron 75:155–160. https://doi.org/10.1016/j.bios.2015.08.034
Tan YN, Su X, Zhu Y, Lee JY (2010) Sensing of transcription factor through controlled-assembly of metal nanoparticles modified with segmented DNA elements. ACS Nano 4(9):5101–5110. https://doi.org/10.1021/nn100943d
Li B, Chen Y, Wang J, Lu Q, Zhu W, Luo J et al (2019) Detecting transcription factors with allosteric DNA-silver nanocluster switches. Anal Chim Acta 1048:168–177. https://doi.org/10.1016/j.aca.2018.10.023
Cao A, Zhang C-y (2013) Real-time detection of transcription factors using target-converted helicase-dependent amplification assay with zero-background signal. Anal Chem 85(4):2543–7. https://doi.org/10.1021/ac400010r
Zhang Y, Hu J, Zhang C-y (2012) Sensitive detection of transcription factors by isothermal exponential amplification-based colorimetric assay. Anal Chem 84(21):9544–9. https://doi.org/10.1021/ac3024087
Deng K, Li C, Huang H, Li X (2017) Rolling circle amplification based on signal-enhanced electrochemical DNA sensor for ultrasensitive transcription factor detection. Sens Actuators B Chem 238:1302–1308. https://doi.org/10.1016/j.snb.2016.09.107
Li C, Qiu X, Hou Z, Deng K (2015) A dumbell probe-mediated rolling circle amplification strategy for highly sensitive transcription factor detection. Biosens Bioelectron 64:505–510. https://doi.org/10.1016/j.bios.2014.09.068
Xu X, Wang L, Zhu D, Wang Y, Jiang W (2018) Protein binding protection in combination with DNA masking for sensitive and reliable transcription factor detection. Talanta 186:293–298. https://doi.org/10.1016/j.talanta.2018.04.047
Zhang K, Wang K, Zhu X, Xie M, Zhang X (2017) A new method for sensitive detection of microphthalmia-associated transcription factor based on “OFF-state” and “ON-state” equilibrium of a well-designed probe and duplex-specific nuclease signal amplification. Biosens Bioelectron 87:299–304. https://doi.org/10.1016/j.bios.2016.08.070
Acknowledgements
We would like to thank the Startup Foundation for Introducing Talent of NUIST (2023r129), the National Natural Science Foundation of China (21964018, 82360259), the Natural Science Foundation of Guangxi Province (2019GXNSFDA245034), and the Guangxi key laboratory of basic and translational research of Bone and joint Degenerative Disease (21-220-06-202205).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval
This research did not involve human or animal samples.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: In this article the affiliation details for Xianjiu Liao were incorrectly given as 'West Guangxi Key Laboratory for Prevention and Treatment of High-Incidence Diseases, Baise 533000, Guangxi, China' but should have been 'West Guangxi Key Laboratory for Prevention and Treatment of High-Incidence Diseases, Youjiang Medical University for Nationalities, Guangxi, Baise, 533000, China'.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, Q., Zhang, C., Deng, X. et al. A CRISPR-Cas12a-based electrochemical biosensor for the detection of microphthalmia-associated transcription factor. Microchim Acta 191, 73 (2024). https://doi.org/10.1007/s00604-023-06164-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-023-06164-5