Abstract
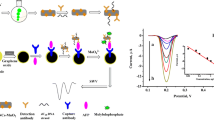
A method is described for the determination of DNA via nucleic acid amplification by using nucleic acid concatemers that result from DNA supersandwich self-assemblies (SSAs). The method employs two auxiliary probes to form self-assembled biotin SSAs. These exhibit strong fluorescence if labeled with intercalator SYBR Green I. In the presence of the target (as exemplified for a 30-mer), streptavidin is released from the surface of the functionalized magnetic microparticles (FMMPs) by competitive hybridization on the surface. However, the SSA products do not conjugate to the FMMPs. This leads to a large amount of SYBR Green I intercalated into the concatemers and eventually results in amplified fluorescence in the supernate. The SSA products can be prepared beforehand, and amplification therefore can be completed within 50 min. The method is more efficient than any other conventional amplification. The detection limit for the 30-mer is 26.4 fM which is better by a factor of 10 compared to other amplification methods. Conceivably, the method can be further extended to the determination of a wide variety of targets simply by replacing the sequences of the probes. Finally, this rapid and highly sensitive method was employed for detection of Ebola virus gene (≈30-mer) and ATP in spiked serum with satisfactory results.

A high sensitivity and efficiency bioassay is described based on functionalized magnetic microparticles and DNA supersandwich self-assemblies.






Similar content being viewed by others
References
Wang F, Lu CH, Willner I (2017) From cascaded catalytic nucleic acids to enzyme–DNA nanostructures: controlling reactivity, sensing, logic operations, and assembly of complex structures. Chem Rev 114:2881–2941
Yuan Z, Zhou Y, Gao S, Cheng Y, Li Z (2014) Homogeneous and sensitive detection of microRNA with ligase chain reaction and lambda exonuclease-assisted cationic conjugated polymer biosensing. ACS Appl Mater Interfaces 6:6181–6185
Yao Q, Wang Y, Wang J, Chen S, Liu H, Jiang Z, Zhang X, Liu SM, Yuan Q, Zhou X (2018) An ultrasensitive diagnostic biochip based on biomimetic periodic nanostructure-assisted rolling circle amplification. ACS Nano 12:6777–6783
Zuo X, Xia F, Xiao Y, Plaxco KW (2010) Sensitive and selective amplified fluorescence DNA detection based on exonuclease III-aided target recycling. J Am Chem Soc 132:1816–1818
Fan D, Zhu X, Zhai Q, Wang E, Dong S (2016) Polydopamine nanotubes as an effective fluorescent quencher for highly sensitive and selective detection of biomolecules assisted with exonuclease III amplification. Anal Chem 88:9158–9165
Peng X, Liang WB, Wen ZB, Xiong CY, Zheng YN, Chai Y, Yuan R (2018) Ultrasensitive fluorescent assay based on a rolling-circle-amplification-assisted multisite-strand-displacement-reaction signal-amplification strategy. Anal Chem 90:7474–7479
Guo Q, Yang X, Wang K, Tan W, Li W, Tang H, Li H (2009) Sensitive fluorescence detection of nucleic acids based on isothermal circular strand-displacement polymerization reaction. Nucleic Acids Res 37:20–25
Ma F, Liu M, Tang B, Zhang C (2017) Rapid and sensitive quantification of microRNAs by isothermal helicase-dependent amplification. Anal Chem 89:6182–6187
Barreda-García S, González-Álvarez MJ, Delos Santos-Álvarez N, Palacios-Gutiérrez JJ, Miranda-Ordieres AJ, Lobo-Castañón MJ (2015) Attomolar quantitation of mycobacterium tuberculosis by asymmetric helicase-dependent isothermal DNA-amplification and electrochemical detection. Biosens Bioelectron 68:122–128
Mudiyanselage APKKK, Yu Q, Leon-Duque MA, Zhao B, Wu R, You M (2018) Genetically encoded catalytic hairpin assembly for sensitive RNA imaging in live cells. J Am Chem Soc 140:8739–8745
Liu C, Chen C, Li S, Dong H, Dai W, Xu T, Liu Y, Yang F, Zhang X (2018)Target-triggered catalytic hairpin assembly-induced core−satellite nanostructures for high-sensitive “off-to-on” SERS detection of intracellular microRNA. Anal Chem 90:10591–10599
Dirks RM, Pierce NA (2004) Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A 101:15275–15278
Huang J, Wang H, Yang X, Quan K, Yang Y, Ying L, Xie N, Ou M, Wang K (2016) Fluorescence resonance energy transfer-based hybridization chain reaction for in situ visualization of tumor-related mRNA. Chem Sci 7:3829–3835
Liu N, Jiang Y, Zhou Y, Xia F, Guo W, Jiang L (2013)Two-way nanopore sensing of sequence-specific oligonucleotides and small-molecule targets in complex matrices using integrated DNA supersandwich structures. Angew Chem Int Ed 52:2007–2011
Jiang Y, Liu N, Guo W, Xia F, Jiang L (2012)Highly-efficient gating of solid-state nanochannels by DNA supersandwich structure containing ATP aptamers: a nanofluidic IMPLICATION logic device. J Am Chem Soc 134:15395–15401
Xia F, White RJ, Zuo X, Patterson A, Xiao Y, Kang D, Gong X, Plaxco KW, Heeger AJ (2010) An electrochemical supersandwich assay for sensitive and selective DNA detection in complex matrices. J Am Chem Soc 132:14346–14348
Xu L, Shen X, Li B, Zhu C, Zhou X (2017)G-quadruplex based Exo III-assisted signal amplification aptasensor for the colorimetric detection of adenosine. Anal Chim Acta 980:58–64
Miao P, Tang Y, Wang B, Yin J, Ning L (2015) Signal amplification by enzymatic tools for nucleic acids. Trends Anal Chem 67:1–15
Gerasimova YV, Kolpashchikov DM (2014)Enzyme-assisted target recycling (EATR) for nucleic acid detection. Chem Soc Rev 43:6405–6438
Zhao Y, Chen F, Li Q, Wang L, Fan C (2015) Isothermal amplification of nucleic acids. Chem Rev 115:12491–12545
Xia Y, Zhang R, Wang Z, Tian J, Chen X (2017) Recent advances in high-performance fluorescent and bioluminescent RNA imaging probes. Chem Soc Rev 46:2824–2843
Zhu Y, Wang H, Wang L, Zhu J, Jiang W (2016) Cascade signal amplification based on copper nanoparticle-reported rolling circle amplification for ultrasensitive electrochemical detection of the prostate cancer biomarker. ACS Appl Mater Interfaces 8:2573–2581
Li C, Wang H, Shen J, Tang B (2015) Cyclometalated iridium complex-based label-free photoelectrochemical biosensor for DNA detection by hybridization chain reaction amplification. Anal Chem 87:4283–4291
Zheng J, Li N, Li C, Wang X, Liu Y, Mao G, Ji X, He Z (2018) A nonenzymatic DNA nanomachine for biomolecular detection by target recycling of hairpin DNA cascade amplification. Biosens Bioelectron 107:40–46
Liu S, Cheng C, Gong H, Wang L (2015) Programmable Mg2+-dependent DNAzyme switch by the catalytic hairpin DNA assembly for dual-signal amplification toward homogeneous analysis of protein and DNA. Chem Commun 51:7364–7367
Yin BC, Ma JL, Le HN, Wang S, Xu Z, Ye BC (2014) A new mode to light up an adjacent DNA-scaffolded silver probe pair and its application for specific DNA detection. Chem Commun 50:15991–15994
Ren W, Liu H, Yang W, Fan Y, Yang L, Wang Y, Liu C, Li Z (2013) A cytometric bead assay for sensitive DNA detection based on enzyme-free signal amplification of hybridization chain reaction. Biosens Bioelectron 49:380–386
Wang S, Yang F, Jin D, Dai Q, Tu J, Liu Y, Ning Y, Zhang GJ (2017) Toehold mediated one-step conformation-switchable “signal-on” electrochemical DNA sensing enhanced with homogeneous enzymatic amplification. Anal Chem 89:5349–5356
Peng Y, Li D, Yuan R, Xiang Y (2018) A catalytic and dual recycling amplification ATP sensor based on target-driven allosteric structure switching of aptamer beacons. Biosens Bioelectron 105:1–5
Li X, Yang J, Xie J, Jiang B, Yuan R, Xiang Y (2018) Cascaded signal amplification via target-triggered formation of aptazyme for sensitive electrochemical detection of ATP. Biosens Bioelectron 102:296–300
Li X, Peng Y, Chai Y, Yuan R, Xiang Y (2016) A target responsive aptamer machine for label-free and sensitive non-enzymatic recycling amplification detection of ATP. Chem Commun 52:3673–3676
Zhang T, Peng Y, Yuan R, Xiang Y (2018)Target-catalyzed assembly formation of metal-ion dependent DNAzymes for non-enzymatic and label-free amplified ATP detection. Sensors Actuators B Chem 273:70–75
Li W, Sun J, Wang H, Wang L, Jiang W (2018) Multifunctional aptamer probe mediated cascade amplification for label-free detection of adenosine. Sensors Actuators B Chem 260:581–586
Song W, Zhang Q, Xie X, Zhang S (2014) Fluorescence aptameric sensor for isothermal circular strand-displacement polymerization amplification detection of adenosine triphosphate. Biosens Bioelectron 61:51–56
Acknowledgements
This work was supported by the National Natural Science Foundation of China (21505114) and Xinxiang Innovative Technology Team (CXTD17004).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 128 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Shangguan, J., He, N. et al. Fluorometric determination of ssDNA based on functionalized magnetic microparticles and DNA supersandwich self-assemblies. Microchim Acta 186, 707 (2019). https://doi.org/10.1007/s00604-019-3865-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3865-z