Abstract
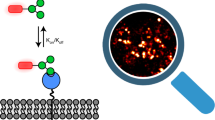
A rolling-mediated cascade (RMC) amplification strategy is described for improved visualization of profiling glycans of mucin 1 (MUC 1) on cell surfaces. CdTe quantum dots (QDs) are used as fluorescent labels. The RMC based amplification allows even distinct glycoforms of MUC1 to be visualized on the surface of MCF-7 cell via an amplified Förster resonance energy transfer (FRET) imaging strategy that works at excitation/emission wavelengths of 345/610 nm. This is achieved by utilizing antibody against MUC1 modified with the fluorescent label 7-amino-4-methylcoumarin-3-acetic acid (AMCA) as the energy donor in FRET. The QDs (used to label surface glycans) act as acceptors. N-Azidoacetylgalactosamine-Acetylated (Ac4GalNAz) as a non-natural azido sugar, can be incorporated into the glycans of the cell surface, which can promote further labeling. The method has the advantage of only requiring a small amount of non-natural sugar to be introduced in metabolic glycan labeling since too much of an artificial sugar will interfere with the physiological functions of cells.

Schematic for the DNA rolling-mediated cascade (RMC)-assisted metabolic labeling of cell surface glycans by using CdTe quantum dots as labels and an intramolecular amplified FRET strategy for imaging glycans on a specific glycosylated protein, MUC1.





Similar content being viewed by others
References
Stowell SR, Ju T, Cummings RD (2015) Protein glycosylation in cancer. Annu Rev Pathol 10(1):473–510. https://doi.org/10.1146/annurev-pathol-012414-040438
Jiang H, English BP, Hazan RB, Wu P, Ovryn B (2015) Tracking surface glycans on live cancer cells with single-molecule sensitivity. Angew Chem Int Ed 127(6):1785–1789. https://doi.org/10.1002/anie.201407976
Paszek MJ, Dufort CC, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, Rubashkin MG, Magbanua MJ, Thorn KS, Davidson MW, Rugo HS, Park JW, Hammer DA, Giannone G, Bertozzi CR, Weaver VM (2014) The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature 511(7509):319–325. https://doi.org/10.1038/nature13535
Wolfert MA, Boons GJ (2013) Adaptive immune activation: Glycosylation does matter. Nat Chem Biol 9(12):776–784. https://doi.org/10.1038/nchembio.1403
Hudak JE, Canham SM, Bertozzi CR (2014) Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol 10(1):69–75. https://doi.org/10.1038/nchembio.1388
Katorcha E, Makarava N, Savtchenko R, Baskakov IV (2014) Sialylation of prion protein controls the rate of prion amplification, the cross-species barrier, the ratio of PrPSc glycoform and prion infectivity. PLoS Pathog 10:e1004366. https://doi.org/10.1371/journal.ppat.1004366
Jaeken J (2013) Congenital disorders of glycosylation. Handb Clin Neurol 113:1737–1743. https://doi.org/10.1038/ejhg.2013.168
Doll F, Buntz A, Spate AK, Schart VF, Timper A, Schrimpf W, Hauck CR, Zumbusch A, Wittmann V (2016) Visualization of protein-specific glycosylation inside living cells. Angew Chem Int Ed 55(6):2262–2266. https://doi.org/10.1002/anie.201503183
Wu N, Bao L, Ding L, Ju H (2016) A single excitation-duplexed imaging strategy for profiling cell surface protein-specific glycoforms. Angew Chem Int Ed 55(17):5220–5224. https://doi.org/10.1002/ange.201601233
Lin W, Du Y, Zhu Y, Chen X (2014) A cis-membrane FRET-based method for protein-specific imaging of cell-surface glycans. J Am Chem Soc 136(2):679–687. https://doi.org/10.1021/ja410086d
Thaysenandersen M, Packer NH (2014) Advances in LC-MS/MS-based glycoproteomics: getting closer to system-wide site-specific mapping of the N- and O-glycoproteome. BBA-Proteins and Proteomics 1844(9):1437–1452. https://doi.org/10.1016/j.bbapap.2014.05.002
Darula Z, Sherman J, Medzihradszky KF (2012) How to dig deeper? Improved enrichment methods for mucin core-1 type glycopeptides. Mol Cell Proteomics 11(7):O111.016774. https://doi.org/10.1074/mcp.O111.016774
Zhang Y, Zhao Y, Ying WT, Qian XH (2018) Progress of O-glycoprotein and O-glycoproteome analysis in secretion systems. Sci Sinica 48(2):124–133. https://doi.org/10.1360/N052017-00170
Kayser H, Zeitler R, Kannicht C, Grunow D, Nuck R, Reutter W (1992) Biosynthesis of a nonphysiological sialic acid in different rat organs, using N-propanoyl-D-hexosamines as precursors. J Biol Chem 267:16934–16938
Robinson PV, de Almeida-Escobedo G, de Groot AE, McKechnie JL, Bertozzi CR (2015) Live-cell labeling of specific protein glycoforms by proximity-enhanced bioorthogonal ligation. J Am Chem Soc 10452-10455. https://doi.org/10.1021/jacs.5b04279
Saxon E, Bertozzi CR (2000) Cell surface engineering by a modified Staudinger reaction. Science 287(5460):2007–2010. https://doi.org/10.1126/science.287.5460.2007
Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc 130(35):11576–11577. https://doi.org/10.1021/ja8030467
Haga Y, Ishii K, Hibino K, Sako Y, Ito Y, Taniguchi N, Suzuki T (2012) Visualizing specific protein glycoforms by transmembrane fluorescence resonance energy transfer. Nat Commun 3(2):907. https://doi.org/10.1038/ncomms1906
Belardi B, de la Zerda A, Spiciarich DR, Maund SL, Peehl DM, Bertozzi CR (2013) Imaging the glycosylation state of cell surface glycoproteins by two-photon fluorescence lifetime imaging. Angew Chem Int Ed 52(52):14045–14049. https://doi.org/10.1002/anie.201307512
Zhang XR, Li R, Chen YY, Zhang SS, Wang WS, Li FC (2016) Applying DNA rolling circle amplification in fluorescence imaging of cell surface glycans labeled by a metabolic method. Chem Sci 7:6182–6189. https://doi.org/10.1039/C6SC02089E
Hui JJ, Bao L, Li SQ, Zhang Y, Feng YM, Ding L, Ju HX (2017) Localized chemical remodeling for live cell imaging of protein-specific glycoform. Angew Chem Int Ed 56:28. https://doi.org/10.1002/ange.201703406
Zhao TB, Li TL, Liu Y (2017) Silver nanoparticle plasmonic enhanced förster resonance energy transfer (FRET) imaging of protein-specific sialylation on the cell surface. Nanoscale 9:9841–9847. https://doi.org/10.1039/C7NR01562C
Jalink K (2013) hiFRET: some tailwind for FRET resolves weak protein interactions. Nat Methods 10(10):947–948. https://doi.org/10.1038/nmeth.2652
Grünberg R, Burnier JV, Ferrar T, Beltran-Sastre V, Stricher F, Sloot AMVD, Garcia-Olivas R, Mallabiabarrena A, Sanjuan X, Zimmermann T, Serrano L (2013) Engineering of weak helper interactions for high-efficiency FRET probes. Nature Method 10(10):1021–1027. https://doi.org/10.1038/NMETH.2625
Chang PV, Chen X, Smyrniotis C, Xenakis A, Hu T, Bertozzi CR, Wu P (2009) Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew Chem Int Ed 121(22):4090–4093. https://doi.org/10.1002/ange.200806319
Laughlin ST, Bertozzi CR (2007) Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat Protoc 2(11):2930–2944. https://doi.org/10.1038/nprot.2007.422
Nath S, Mukherjee P (2014) MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol Med 20(6):332–342. https://doi.org/10.1016/j.molmed.2014.02.007
Zhang S, Wen L, Yang J, Zeng J, Sun Q (2016) Facile fabrication of dendritic Mesoporous SiO2@CdTe@SiO2 fluorescent nanoparticles for bioimaging. Part Part Syst Charact 33(5):261–270. https://doi.org/10.1002/ppsc.201500254
Zhu D, Miao ZY, Hu Y, Zhang XJ (2018) Single-step, homogeneous and sensitive detection for microRNAs with dual recognition steps based on luminescence resonance energy transfer (LRET) using upconversion nanoparticles. Biosens Bioelectron 100:475–481. https://doi.org/10.1016/j.bios.2017.09.039
Michalet X, Pinaud FF, Bentolila A, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S (2005) Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307:538–544. https://doi.org/10.1126/science.1104274
Rong J, Han J, Dong L, Tan Y, Yang H, Feng L, Wang Q, Meng R, Zhao J, Wang S, Chen X (2014) Glycan imaging in intact rat hearts and glycoproteomic analysis reveal the upregulation of sialylation during cardiac hypertrophy. J Am Chem Soc 136(50):17468–17476. https://doi.org/10.1021/ja508484c
Acknowledgments
We sincerely appreciate the National Natural Science Foundation of China for the financial support (81573388). This work was supported by “Qing Lan Project of Jiangsu province” and “Six talent peaks project of Jiangsu Province (YY-032)”. This work was also supported by the Open Project Program of Jiangsu Key Laboratory for Pharmacology and Safety Evaluation of Chinese Materia Medica (No. JKLPSE201805) and the Project of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 5142 kb)
Rights and permissions
About this article
Cite this article
Yang, X., Tang, Y., Zhang, X. et al. Fluorometric visualization of mucin 1 glycans on cell surfaces based on rolling-mediated cascade amplification and CdTe quantum dots. Microchim Acta 186, 721 (2019). https://doi.org/10.1007/s00604-019-3840-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00604-019-3840-8