Abstract
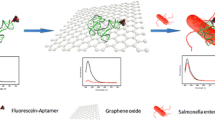
This article describes a sensitive and selective fluorometric method for the determination of Salmonella enteritidis by exploiting the polymerase activity of the Klenow fragment and dual fluorescence. First, one end of a target-selective aptamer was labeled with the fluorophore 6-carboxyfluorescein (FAM). Once the labeled aptamer binds to graphene oxide (GO) via π-stacking interaction, the fluorescence of FAM is quenched. However, the addition of target (16S rRNA) leads to the restoration of fluorescence due to the binding of probe and target which shifts the FAM fluorophore away from the quenching GO. By using the Klenow fragment and by exploiting the synergistic effect of FAM and the DNA probe SYBR Green I (which is strongly fluorescent in presence of dsDNA only), fluorescence is strongly amplified and sensitivity improved. The analyte 16SrRNA can be determined by this method in the 60 pM to 100 nM concentration range, and the detection limit is 60 pM. It is also shown that Salmonella enteritidis can be determined in milk samples by this method in concentrations between 102 to 105 cfu⋅mL‾1, with a detection limit of 300 cfu⋅mL‾1. This assay displays high sensitivity and selectivity and may possess wide applications in pathogen detection.

If labeled aptamer against Salmonella enteritidis 16S rRNA binds to graphene oxide GO) by π-interaction, the fluorescence of the label is quenched. The addition of target rRNA leads to the restoration of fluorescence, and this effect can be used to quantify Salmonella enteritidis.





Similar content being viewed by others
References
Rodrigue DC, Tauxe RV, Rowe B (1990) International increase in salmonella enteritidis: a new pandemic? Epidemiol Infect 105:21–27
Pui CF, Wong WC, Chai LC, Tunung R, Jeyaletchumi P, Noor Hidayah MS, Ubong A, Farinazleen MG, Cheah YK, Son R (2011) Salmonella: a foodborne pathogen. International Food Research Journal 18:465–473
Herikstad H, Motarjemi Y, Tauxe RV (2002) Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect 129:1–8
Allen MJ, Edberg SC, Reasoner DJ (2004) Heterotrophic plate count bacteria-what is their significance in drinking water? Int J Food Microbiol 92(3):265–274
Uyttendaele M, Vanwildemeersch K, Debevere J (2003) Evaluation of real-time PCR vs automated ELISA and a conventional culture method using a semi-solid medium for detection of salmonella. Lett Appl Microbiol 37:386–391
Martin B, Garriga M, Aymerich T (2012) Pre-PCR treatments as a key factor on the probability of detection of Listeria monocytogenes and salmonella in readyto-eat meat products by real-time PCR. Food Control 27(1):163–169
Sanvicens N, Pastells C, Pascual N, Marco MP (2009) Nanoparticle-based aptasensors for detection of pathogenic bacteria. Trends Anal Chem 28(11):1243–1252
Sanvicens N, Pascual N, Fernández-Argüelles MT, Adrián J, Costa-Fernández JM, Sánchez-Baeza F, et al. (2011) Quantum dot-based array for sensitive detection of Escherichia coli. Anal Bioanal Chem 399(8):2755–2762
Lungu B, Waltman WD, Berghaus RD, Hofacre CL (2012) Comparison of a real-time PCR method with a culture method for the detection of Salmonella enterica serotype enteritidis in naturally contaminated environmental samples from integrated poultry houses. J Food Prot Apr. 75(4):743–747. doi:10.4315/0362-028X.JFP-11-297
Sokolov DM, Sokolov MS (2013) Rapid methods for the genus salmonella bacteria detection in food and raw materials. Vopr Pitan 82:33–40
Zhi Y, Sasai D, Okubo Y, Shinozaki M, Nakayama H, Yamagata Murayama S, Wakayama M, Ide T, Zhang Z, Shibuya K (2013) Comparison between the effectiveness of polymerase chain reaction and in situ hybridization in detecting the presence of pathogenic fungi by using the preserved DNA in formalin-fixed and paraffin-embedded tissues. Jpn J Infect Dis 66:173–179
Lei P, Tang H, Ding S, et al. (2015) Determination of the invA gene of salmonella using surface Plasmon resonance along with streptavidin aptamer amplification. Microchim Acta 182:289–296
Jia F, Duan N, Wu S, Dai R, Wang Z, Li X (2015) Impedimetric salmonella aptasensor using a glassy carbon electrode modified with an electrodeposited composite consisting of reduced graphene oxide and carbon nanotubes. Microchim Acta. doi:10.1007/s00604-015-1649-7
Fei J, Dou W, Zhao G (2015) A sandwich electrochemical immunosensor for salmonella pullorum and salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Microchim Acta. doi:10.1007/s00604-015-1573-x
Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA (1990) Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol 56:1919–1925
Harvey I, Cormier Y, Beaulieu C, Akimov VN, Meriaux A, Duchaine C (2001) Random amplified ribosomal DNA restriction analysis for rapid identification of thermophilic actinomycete-like bacteria involved in hypersensitivity pneumonitis. Syst Appl Microbiol 24:277–284
Robinson JT, Perkins FK, Snow ES, Wei ZQ, Sheehan PE (2008) Reduced graphene oxide molecular sensors. Nano Lett 8:3137–3140
Wu J, Wang YS, Yang XY, Liu YY, Yang JR, Yang R, Zhang N (2012) Graphene oxide used as a carrier for Adriamycin can reverse drug resistance in breast cancercells. Nanotechnology 23:355101. doi:10.1088/0957-4484/23/35/355101
Yin D, Li Y, Lin H, Guo B, Du Y, Li X, Jia H, Zhao X, Tang J, Zhang L (2013) Functional graphene oxide as a plasmid-based Stat3 siRNA carrier inhibits mouse malignant melanoma growth in vivo. Nanotechnology 24:105102. doi:10.1088/0957-4484/24/10/105102
Pérez-López B, Merkoçi A (2012) Carbon nanotubes and graphene in analytical sciences. Microchim Acta 179:1–16
Duan YF, Ning Y, Song Y, Deng L (2014) Fluorescent aptasensor for the determination of salmonella typhimurium based on a graphene oxide platform. Microchim Acta 181:647–653
Xiang DS, Zhou GH, Luo M, Ji XH, He ZK (2012) Dual color fluorescence quantitative detection of specific single-stranded DNA with molecular beacons and nucleic acid dye SYBR green I. Analyst 137:3787–3793
Cordeiro M, Giestas L, Lima JC, Baptista PJ (2013) Coupling an universal primer to SBE combined spectral codification strategy for single nucleotide polymorphism analysis. Biotechnol. 168(1):90–94
Singer VL, Lawlor TE, Yue S (1999) Comparison of SYBR green I nucleic acid gel stain mutagenicity and ethidium bromide mutagenicity in the salmonella/mammalian microsome reverse mutation assay (ames test). Mutat Res 439(1):37–47
Xiang D, Zhai K, Xiang W, Wang L (2014) Highly sensitive fluorescence quantitative detection of specific DNA sequences with molecular beacons and nucleic acid dye SYBR green I. Talanta 129:249–253
Song Y, Li WK, Duan YF, Li ZJ, Deng L (2014) Nicking enzyme-assisted aptasensor for salmonella enteritidis detection based on fluorescence resonance energy transfer. Biosens Bioelectron 55:400–404
Li S, Li Y, Chen H, Horikawa S, Shen W, Simonian A, Chin BA (2010) Direct detection of salmonella typhimurium on fresh produce using phage-based magnetoelastic aptasensors. Biosens Bioelectron 26:1313–1319
Ohk S-H, Bhunia AK (2013) Multiplex fiber optic aptasensor for detection of Listeria monocytogenes, Escherichia coli O157:H7 and Salmonella enterica from ready-to-eat meat samples. Food Microbiol 33:166–171
Duan N, Wu S, Yu Y, Ma X, Xia Y, Chen X, Huang Y, Wang Z (2013) A dual-color flow cytometry protocol for the simultaneous detection of vibrio parahaemolyticus and salmonella typhimurium using aptamer conjugated quantum dots as labels. Anal Chim Acta 804:151–158
Wu X, Xu C, Tripp RA, Huang YW, Zhao Y (2013) Detection and differentiation of foodborne pathogenic bacteria in mung bean sprouts using field deployable label-free SERS devices. Analyst 138:3005–3012
Wu WH, Li M, Wang Y, Ouyang HX, Wang L, Li CX, Cao YC, Meng QH, Lu JX (2012) Aptasensors for rapid detection of Escherichia coli O157:H7 and salmonella typhimurium. Nanoscale Res Lett 7:658
Nguyen P-D, Tran TB, Nguyen DTX, et al. (2014) Magnetic silica nanotube-assisted impedimetric immunosensor for the separation and label-free detection of salmonella typhimurium. Sensors Actuators B Chem 197:314–320
Wu W, Li J, Pan D, Li J, Song S, RongM LZ, Gao J, Lu J (2014) Gold nanoparticle-based enzyme-linked antibody-aptamer sandwich assay for detection of salmonella typhimurium. ACS Appl Mater Interfaces 6(19):16974–16981
Acknowledgments
This work was financially supported by the National Natural Science Foundation (81271660), Doctoral Program of Higher Education from the Ministry of Education (20114306110006).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 305 kb)
Rights and permissions
About this article
Cite this article
Liu, K., Yan, X., Mao, B. et al. Aptamer-based detection of Salmonella enteritidis using double signal amplification by Klenow fragment and dual fluorescence. Microchim Acta 183, 643–649 (2016). https://doi.org/10.1007/s00604-015-1692-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00604-015-1692-4