Abstract
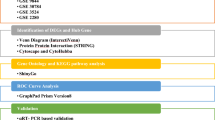
Cancer is one of the world’s most deadly diseases with high economic implications. Early detection can increase the survival chance of the patient, but at present, it remains a challenge due to the non-specificity or absence of symptoms at the onset. The occurrence of multiple cancer types in a single individual is increasingly being reported especially among cancer survivors. This study examines the potential of salivary biomarkers in early detection of one or more cancer types. Four gene expression datasets of salivary biomarkers in pancreatic, lung, ovarian, and breast cancers (GSE53325, GSE29220, GSE32175, and GSE20266, respectively) were examined, and their differentially expressed genes (DEGs) were determined using GEO2R. The commonly occurring DEGs were determined using Venny and were functionally characterized using the Database for Annotation, Visualization and Integrated Discovery (DAVID). The study identified 159 commonly upregulated genes in breast and lung cancers, 29,453 commonly downregulated in “Lung” and “Ovarian” cancers, 1489 genes were commonly downregulated in breast and ovarian cancers, and 16,612 genes were commonly downregulated in lung, breast, and ovarian cancers, while only a gene was found to be commonly downregulated in lung and breast cancer. The protein–protein interactions network revealed two key genes (UBB and FN1) that are centrally linked to others in commonly upregulated breast and lung cancers. Therefore, the commonly DEGs could serve as potential biomarkers to the prognosis and diagnosis of the common occurrence of the different selected types of cancer. Also, UBB and FN1 are key biomarkers for the co-occurrence of breast and lung cancers.










Similar content being viewed by others
References
Azameti MK, Dauda WP, Panzade KP, Vishwakarma H (2021) 2021) Identification and characterization of genes responsive to drought and heat stress in rice (Oryza sativa L. Vegetos 34(2):309–317. https://doi.org/10.1007/s42535-021-00198-x
Bacильeв CA, Гopгидзe Л A, Eфpeмoв EE, Бeлинин ГЮ, Moиceeвa TH, Coкoлoвa MA, Vasiliev SA, Gorgidze LA, Efremov EE, Belinin GY, Moiseeva, TN (2022) клиничecкaя знaчимocть ( oбзop ) Fibronectin : structure , functions , clinical significance ( review ). 12(1):138–158.
Bui QT, Hong JH, Kwak M, Lee JY, Lee PCW (2021) Ubiquitin-conjugating enzymes in cancer. In Cells 10(6):1–16. https://doi.org/10.3390/cells10061383
Cheng SS, Yang GJ, Wang W, Leung CH, Ma DL (2020) The design and development of covalent protein-protein interaction inhibitors for cancer treatment. J Hematol Oncol 13:1–14
Chhikara BS, Parang K (2023) Global Cancer Statistics 2022: the trends projection analysis. Chem Biol Lett 10(1):1–16
Deng H, Huang Y, Wang L, Chen M (2020) High expression of UBB, Rac1, and ITGB1 predicts worse prognosis among nonsmoking patients with lung adenocarcinoma through bioinformatics analysis. BioMed Research International, 2020. https://doi.org/10.1155/2020/2071593
Kanchan V, Navin V, Ajay R, Basabi R (2020) Transcription factors in cancer development and therapy. Cancers 12(2296):725–729
Kar G, Gursoy A, Keskin O (2009) Human cancer protein-protein interaction network: a structural perspective. PLoS Computational Biology, 5(12). https://doi.org/10.1371/journal.pcbi.1000601
Kurian T (2024) In silico screening by molecular docking of heterocyclic compounds with furan or indole nucleus from database for anticancer activity and validation of the method by redocking. 16(4):4–7
Lee YH, Kim JH, Zhou H, Kim BW, Wong DT (2012) Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J Mol Med 90(4):427–434. https://doi.org/10.1007/s00109-011-0829-0
Li K, Lin Y, Zhou Y, Xiong X, Wang L, Li J, Zhou F, Guo Y, Chen S, Chen Y, Tang H, Qiu X, Cai S (2023) Salivary extracellular MicroRNAs for early detection and prognostication of esophageal cancer : a clinical study. Gastroenterology 165(4):932–945.e9. https://doi.org/10.1053/j.gastro.2023.06.021
Rapado-González Ó, Martínez-Reglero C, Salgado-Barreira Á, Takkouche B, López-López R, Suárez-Cunqueiro MM, Muinelo-Romay L (2020) Salivary biomarkers for cancer diagnosis: a meta-analysis. Ann Med 52(3–4):131–144. https://doi.org/10.1080/07853890.2020.1730431
Sun F, Zhou JL, Peng PJ, Qiu C, Cao JR, Peng H (2021) Identification of Disease-Specific Hub Biomarkers and Immune Infiltration in Osteoarthritis and Rheumatoid Arthritis Synovial Tissues by Bioinformatics Analysis. Dis Markers. https://doi.org/10.1155/2021/9911184
Wang X, Kaczor-Urbanowicz KE, Wong DTW (2017) Salivary biomarkers in cancer detection. Med Oncol 34(1):1–8. https://doi.org/10.1007/s12032-016-0863-4
Xie Z, Yin X, Gong B, Nie W, Wu B, Zhang X, Huang J, Zhang P, Zhou Z, Li Z (2015) Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev Res 8(2):165–173. https://doi.org/10.1158/1940-6207.CAPR-14-0192
Yoneda A (2015) Fibronectin matrix assembly and its significant role in cancer progression and treatment. Trends Glycosci Glycotechnol 27(155):89–98. https://doi.org/10.4052/tigg.1421.1
Zamanian-Azodi M, Rezaei-Tavirani M, Rahmati-Rad S, Hasanzadeh H, Tavirani MR, Seyyedi SS (2015) Protein-protein interaction network could reveal the relationship between the breast and colon cancer. Gastroenterol Hepatol Bed to Bench 8(3):215–224
Zhang L, Xiao H, Karlan S, Zhou H, Gross J, Elashoff D, Akin D, Yan X, Chia D, Karlan B, Wong DT (2010) Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non- invasive detection of breast cancer. PLoS One 5(12):1–7. https://doi.org/10.1371/journal.pone.0015573
Zhang L, Xiao H, Zhou H, Santiago S, Lee JM, Garon EB, Yang J, Brinkmann O, Yan X, Akin D, Chia D, Elashoff D, Park NH, Wong DTW (2012) Development of transcriptomic biomarker signature in human saliva to detect lung cancer. Cell Mol Life Sci 69:3341–3350
Zhang XX, Luo JH, Wu LQ (2022) FN1 overexpression is correlated with unfavorable prognosis and immune infiltrates in breast cancer. Front Genet 13(August):1–16. https://doi.org/10.3389/fgene.2022.913659
Zhou WH, Du WD, Li YF, Al-Aroomi MA, Yan C, Wang Y, Zhang ZY, Liu FY, Sun CF (2022) The overexpression of fibronectin 1 promotes cancer progression and associated with M2 macrophages polarization in head and neck squamous cell carcinoma patients. Int J Gen Med 15(May):5027–5042. https://doi.org/10.2147/IJGM.S364708
Zhou Y, Cao G, Cai H, Huang H, Zhu X (2020) The effect and clinical significance of FN1 expression on biological functions of gastric cancer cells. Cell Mol Biol (Noisy-le-grand). 66(5):191–198
Funding
This study was not supported by any funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
For this type of study informed consent is not required.
Consent for publication
For this type of study consent for publication is not required.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Glen, E., Dauda, W.P., Abraham, P. et al. Overexpression of salivary UBB and FN1 genes as potential biomarkers for the prognosis and diagnosis of co-occurrence in breast and lung cancers. Comp Clin Pathol (2025). https://doi.org/10.1007/s00580-025-03662-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00580-025-03662-1