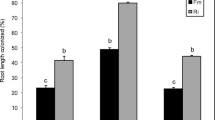
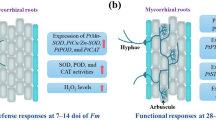
Abstract
Arbuscular mycorrhizal (AM) fungi form a root endosymbiosis with many agronomically important crop species. They enhance the ability of their host to obtain nutrients from the soil and increase the tolerance to biotic and abiotic stressors. However, AM fungal species can differ in the benefits they provide to their host plants. Here, we examined the putative molecular mechanisms involved in the regulation of the physiological response of Medicago truncatula to colonization by Rhizophagus irregularis or Glomus aggregatum, which have previously been characterized as high- and low-benefit AM fungal species, respectively. Colonization with R. irregularis led to greater growth and nutrient uptake than colonization with G. aggregatum. These benefits were linked to an elevated expression in the roots of strigolactone biosynthesis genes (NSP1, NSP2, CCD7, and MAX1a), mycorrhiza-induced phosphate (PT8), ammonium (AMT2;3), and nitrate (NPF4.12) transporters and the putative ammonium transporter NIP1;5. R. irregularis also stimulated the expression of photosynthesis-related genes in the shoot and the upregulation of the sugar transporters SWEET1.2, SWEET3.3, and SWEET 12 and the lipid biosynthesis gene RAM2 in the roots. In contrast, G. aggregatum induced the expression of biotic stress defense response genes in the shoots, and several genes associated with abiotic stress in the roots. This suggests that either the host perceives colonization by G. aggregatum as pathogen attack or that G. aggregatum can prime host defense responses. Our findings highlight molecular mechanisms that host plants may use to regulate their association with high- and low-benefit arbuscular mycorrhizal symbionts.









Similar content being viewed by others
Data and material availability
The RNA-seq datasets generated for and analyzed in the current study are available on NCBI as BioProject ID PRJNA832760 and can be accessed at http://www.ncbi.nlm.nih.gov/bioproject/832760.
References
Aloui A, Recorbet G, Lemaître-Guillier C et al (2018) The plasma membrane proteome of Medicago truncatula roots as modified by arbuscular mycorrhizal symbiosis. Mycorrhiza 28:1–16. https://doi.org/10.1007/s00572-017-0789-5
An J, Zeng T, Ji C et al (2019) A Medicago truncatula SWEET transporter implicated in arbuscule maintenance during arbuscular mycorrhizal symbiosis. New Phytol 224:396–408. https://doi.org/10.1111/nph.15975
Besserer A, Becard G, Jauneau A et al (2008) GR24, a synthetic analog of strigolactones, stimulates the mitosis and growth of the abuscular mycorrhizal fungus Gigaspora rosea by boosting its energy metabolism. Plant Physiol 148:402–413. https://doi.org/10.1104/pp.108.121400
Boddey RM, Polidoro JC, Resende AS et al (2001) Use of the 15N natural abundance technique for the quantification of the contribution of N2 fixation to sugar cane and other grasses. Aust J Plant Physiol 28:889–895. https://doi.org/10.1071/PP01058
Bravo A, Brands M, Wewer V et al (2017) Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytol 214:1631–1645. https://doi.org/10.1111/nph.14533
Breuillin-Sessoms F, Floss DS, Karen Gomez S et al (2015) Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 27:1352–1366. https://doi.org/10.1105/tpc.114.131144
Brundrett MC, Tedersoo L (2018) Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol 220:1108–1115. https://doi.org/10.1111/nph.14976
Bücking H, Kafle A (2015) Role of arbuscular mycorrhizal fungi in the nitrogen uptake of plants: current knowledge and research gaps. Agronomy 5:587–612. https://doi.org/10.3390/agronomy5040587
Casieri L, Gallardo K, Wipf D (2012) Transcriptional response of Medicago truncatula sulphate transporters to arbuscular mycorrhizal symbiosis with and without sulphur stress. Planta 235:1431–1447. https://doi.org/10.1007/s00425-012-1645-7
Chandrasekaran M (2020) A meta-analytical approach on arbuscular mycorrhizal fungi inoculation efficiency on plant growth and nutrient uptake. Agriculture 10:370. https://doi.org/10.3390/agriculture10090370
Chen HY, Huh JH, Yu YC et al (2015) The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J 83:1046–1058. https://doi.org/10.1111/tpj.12948
Cope KR, Bascaules A, Irving TB et al (2019) The ectomycorrhizal fungus Laccaria bicolor produces lipochitooligosaccharides and uses the common symbiosis pathway to colonize populus roots. Plant Cell 31:2386–2410. https://doi.org/10.1105/tpc.18.00676
Davière JM, Achard P (2013) Gibberellin Signaling in Plants Development 140:1147–1151. https://doi.org/10.1242/dev.087650
De Carvalho EX, Menezes RSC, Santiago de Freitas AD et al (2017) The 15N natural abundance technique to assess the potential of biological nitrogen fixation (BNF) in some important C4 grasses. AJCS 11:1559–1564. https://doi.org/10.21475/ajcs.17.11.12.pne729
Doidy J, Vidal U, Lemoine R (2019) Sugar transporters in Fabaceae, featuring SUT MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum. PLoS One 14:e0223173. https://doi.org/10.1371/journal.pone.0223173
Fellbaum CR, Gachomo EW, Beesetty Y et al (2012) Carbon availability triggers fungal nitrogen uptake and transport in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 109:2666–2671. https://doi.org/10.1073/pnas.1118650109
Fellbaum CR, Mensah JA, Cloos AJ et al (2014) Fungal nutrient allocation in common mycorrhizal networks is regulated by the carbon source strength of individual host plants. New Phytol 203:646–656. https://doi.org/10.1111/nph.12827
Feng F, Sun J, Radhakrishnan GV et al (2019) A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat Commun 10:1–12. https://doi.org/10.1038/s41467-019-12999-5
Floss DS, Gomez SK, Park HJ et al (2017) A transcriptional program for arbuscule degeneration during AM symbiosis is regulated by MYB1. Curr Biol 27:1206–1212. https://doi.org/10.1016/j.cub.2017.03.003
Floss DS, Levy JG, Lévesque-Tremblay V et al (2013) DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci U S A 110:E5025–E5034. https://doi.org/10.1073/pnas.1308973110
Frare R, Ayub N, Alleva K, Soto G (2018) The ammonium channel NOD26 is the evolutionary innovation that drives the emergence, consolidation, and dissemination of nitrogen-fixing symbiosis in angiosperms. J Mol Evol 86:554–565. https://doi.org/10.1007/s00239-018-9867-3
Garcia K, Doidy J, Zimmermann SD et al (2016) Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci 21:937–950
Genre A, Chabaud M, Balzergue C et al (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198:190–202. https://doi.org/10.1111/nph.12146
Giovannetti M, Tolosano M, Volpe V et al (2014) Identification and functional characterization of a sulfate transporter induced by both sulfur starvation and mycorrhiza formation in Lotus japonicus. New Phytol 204:609–619. https://doi.org/10.1111/nph.12949
Goff LA, Trapnell C, Kelley D (2012) CummeRbund: visualization and exploration of Cufflinks high-throughput sequencing data. R Packag Version 2:1–45
Grunwald U, Guo W, Fischer K et al (2009) Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 229:1023–1034. https://doi.org/10.1007/s00425-008-0877-z
Hart MM, Forsythe J, Oshowski B et al (2013) Hiding in a crowd - does diversity facilitate persistence of a low-quality fungal partner in the mycorrhizal symbiosis? Symbiosis 59:47–56. https://doi.org/10.1007/s13199-012-0197-8
Havlin JL, Beaton JD, Tisdale SL, Nelson WL (2014) Soil fertility and fertilizers: an introduction to nutrient management. Pearson Education Inc, Eighth
Heck C, Kuhn H, Heidt S et al (2016) Symbiotic fungi control plant root cortex development through the novel GRAS transcription factor MIG1. Curr Biol 26:2770–2778. https://doi.org/10.1016/j.cub.2016.07.059
Horváth B, Yeun LH, Domonkos Á et al (2011) Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol Plant-Microbe Interact 24:1345–1358. https://doi.org/10.1094/mpmi-01-11-0015
Ingestad T (1960) Studies on the nutrition of forest tree seedlings. III mineral nutrition of pine. Physiol Plant 13:513–533. https://doi.org/10.1111/j.1399-3054.1960.tb08074.x
Jansa J, Smith FA, Smith SE (2008) Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol 177:779–789. https://doi.org/10.1111/j.1469-8137.2007.02294.x
Javot H, Penmetsa RV, Terzaghi N et al (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci 104:1720–1725. https://doi.org/10.1073/pnas.0608136104
Johnson NC, Graham JH (2013) The continuum concept remains a useful framework for studying mycorrhizal functioning. Plant Soil 363:411–419. https://doi.org/10.1007/s11104-012-1406-1
Kafle A, Cope KR, Raths R et al (2019) Harnessing soil microbes to improve plant phosphate efficiency in cropping systems. Agronomy 9:127. https://doi.org/10.3390/agronomy9030127
Kafle A, Frank HER, Rose BD, Garcia K (2022) Split down the middle: studying arbuscular mycorrhizal and ectomycorrhizal symbioses using split-root assays. J Exp Bot 73:1288–1300. https://doi.org/10.1093/JXB/ERAB489
Kafle A, Garcia K, Wang X et al (2018) Nutrient demand and fungal access to resources control the carbon allocation to the symbiotic partners in tripartite interactions of Medicago truncatula. Plant Cell Environ 1–15. https://doi.org/10.1111/pce.13359
Kaschuk G, Kuyper TW, Leffelaar PA et al (2009) Are the rates of photosynthesis stimulated by the carbon sink strength of rhizobial and arbuscular mycorrhizal symbioses? Soil Biol Biochem 41:1233–1244. https://doi.org/10.1016/j.soilbio.2009.03.005
Kiers ET, Duhamel M, Beesetty Y et al (2011) Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science (80- ) 333:880–882. https://doi.org/10.1126/science.1208473
Kim D, Paggi JM, Park C et al (2019a) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. https://doi.org/10.1038/s41587-019-0201-4
Kim S, Zeng W, Bernard S et al (2019b) Ca2+-regulated Ca2+ channels with an RCK gating ring control plant symbiotic associations. Nat Commun 10:1–12. https://doi.org/10.1038/s41467-019-11698-5
Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209. https://doi.org/10.1016/j.cub.2011.06.044
Konečný J, Hršelová H, Bukovská P et al (2019) Correlative evidence for co-regulation of phosphorus and carbon exchanges with symbiotic fungus in the arbuscular mycorrhizal Medicago truncatula. PLoS One 14. https://doi.org/10.1371/journal.pone.0224938
Kretzschmar T, Kohlen W, Sasse J et al (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483:341–344. https://doi.org/10.1038/nature10873
LeBauer DS, Treseder KK (2008) Nitrogen limitation of net primary productivity in terrestrial ecosystems is globally distributed. Ecology 89:371–379. https://doi.org/10.1890/06-2057.1
Lendenmann M, Thonar C, Barnard RL et al (2011) Symbiont identity matters: carbon and phosphorus fluxes between Medicago truncatula and different arbuscular mycorrhizal fungi. Mycorrhiza 21:689–702. https://doi.org/10.1007/S00572-011-0371-5/FIGURES/4
Lévy J, Bres C, Geurts R et al (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science (80- ) 303:1361–4. https://doi.org/10.1126/science.1093038
Li Y, Duan T, Nan Z, Li Y (2019a) Arbuscular mycorrhizal fungus alleviates alfalfa leaf spots caused by Phoma medicaginis revealed by RNA-seq analysis. J Appl Microbiol. https://doi.org/10.1111/jam.14387
Li Y, Nan Z, Duan T (2019b) Rhizophagus intraradices promotes alfalfa (Medicago sativaM) defense against pea aphids (Acyrthosiphon pisum) revealed by RNA-Seq analysis. Mycorrhiza 29:623–635. https://doi.org/10.1007/s00572-019-00915-0
Liu J, Maldonado-Mendoza I, Lopez-Meyer M et al (2007) Arbuscular mycorrhizal symbiosis is accompanied by local and systemic alterations in gene expression and an increase in disease resistance in the shoots. Plant J 50:529–544. https://doi.org/10.1111/j.1365-313X.2007.03069.x
MacLean AM, Bravo A, Harrison MJ (2017) Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell 29:2319–2335. https://doi.org/10.1105/tpc.17.00555
Maillet F, Poinsot V, André O et al (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469:58–64. https://doi.org/10.1038/nature09622
Maróti G, Downie JA, Kondorosi É (2015) Plant cysteine-rich peptides that inhibit pathogen growth and control rhizobial differentiation in legume nodules. Curr Opin Plant Biol 26:57–63. https://doi.org/10.1016/j.pbi.2015.05.031
Martin FM, Uroz S, Barker DG (2017) Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science (80-) 356:eaad4501
McGonigle TP, Miller MH, Evans DG et al (1990) A new method which gives an objective measure of colonization of roots by vesicular-arbuscular mycorrhizal fungi. New Phytol 115:495–501. https://doi.org/10.1111/j.1469-8137.1990.tb00476.x
Müller LM, Flokova K, Schnabel E et al (2019) A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nat Plants 5:933–939. https://doi.org/10.1038/s41477-019-0501-1
Nadal M, Sawers R, Naseem S et al (2017) An N-acetylglucosamine transporter required for arbuscular mycorrhizal symbioses in rice and maize. Nat Plants 3:1–7. https://doi.org/10.1038/nplants.2017.73
Nouri E, Surve R, Bapaume L et al (2021) Phosphate suppression of arbuscular mycorrhizal symbiosis involves gibberellic acid signaling. Plant Cell Physiol. https://doi.org/10.1093/pcp/pcab063
Park HJ, Floss DS, Levesque-Tremblay V et al (2015) Hyphal branching during arbuscule development requires RAM1. Plant Physiol 169. https://doi.org/10.1104/pp.15.01155
Paul EA, Kucey RMN (1981) Carbon flow in plant microbial associations incriminated in perinatal morbidity and mortality. Science (80-) 213:473–474. https://doi.org/10.1126/science.213.4506.473
Pertea M, Pertea GM, Antonescu CM et al (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. https://doi.org/10.1038/nbt.3122
Pimprikar P, Carbonnel S, Paries M et al (2016) A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol 26:987–998. https://doi.org/10.1016/j.cub.2016.01.069
Pimprikar P, Gutjahr C (2018) Transcriptional regulation of arbuscular mycorrhiza development. Plant Cell Physiol 59:673–690
Postnikova OA, Shao J, Nemchinov LG (2013) Analysis of the alfalfa root transcriptome in response to salinity stress. Plant Cell Physiol 54:1041–1055. https://doi.org/10.1093/pcp/pct056
Roth R, Paszkowski U (2017) Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr Opin Plant Biol 39:50–56. https://doi.org/10.1016/j.pbi.2017.05.008
Roux SJ, Clark GB, Morgan RO et al (2012) Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol 196:695–712. https://doi.org/10.1111/j.1469-8137.2012.04308.x
Roy S, Breakspear A, Cousins D et al (2021) Three common symbiotic ABC-B transporters in Medicago truncatula are regulated by a NIN-independent branch of the symbiosis signalling pathway. Mol Plant-Microbe Interact. https://doi.org/10.1094/mpmi-02-21-0036-r
Rush TA, Puech-Pagès V, Bascaules A et al (2020) Lipo-chitooligosaccharides as regulatory signals of fungal growth and development. Nat Commun 11:1–10. https://doi.org/10.1038/s41467-020-17615-5
Sameeullah M, Demiral T, Aslam N et al (2016) In silico functional analyses of SWEETs reveal cues for their role in AMF symbiosis. In: Plant, soil and microbes: volume 2: mechanisms and molecular interactions. Springer International Publishing, pp 45–58
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Sharma E, Anand G, Kapoor R (2017) Terpenoids in plant and arbuscular mycorrhiza-reinforced defence against herbivorous insects. Ann Bot 119:791–801
Sieh D, Watanabe M, Devers EA et al (2013) The arbuscular mycorrhizal symbiosis influences sulfur starvation responses of Medicago truncatula. New Phytol 197:606–616. https://doi.org/10.1111/nph.12034
Singh S, Parniske M (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15:444–453
Spatafora JW, Chang Y, Benny GL et al (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108:1028–1046. https://doi.org/10.3852/16-042
St-Arnaud M, Hamel C, Vimard B et al (1996) Enhanced hyphal growth and spore production of the arbuscular mycorrhizal fungus Glomus intraradices in an in vitro system in the absence of host roots. Mycol Res 100:328–332. https://doi.org/10.1016/S0953-7562(96)80164-X
Stracke S, Kistner C, Yoshida S et al (2002) A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417:959–962. https://doi.org/10.1038/nature00841
Stuurman N, Pacios Bras C, Schlaman HRM et al (2007) Use of green fluorescent protein color variants expressed on stable broad-host-range vectors to visualize rhizobia interacting with plants. 13:1163–1169. https://doi.org/10.1094/MPMI.2000.13.11.1163
Tang H, Krishnakumar V, Bidwell S et al (2014) An improved genome release (version Mt4.0) for the model legume Medicago truncatula. BMC Genomics 15:312. https://doi.org/10.1186/1471-2164-15-312
Tejada-Jiménez M, Gil-Díez P, León-Mediavilla J et al (2017) Medicago truncatula molybdate transporter type 1 (MtMOT1.3) is a plasma membrane molybdenum transporter required for nitrogenase activity in root nodules under molybdenum deficiency. New Phytol 216:1223–1235. https://doi.org/10.1111/nph.14739
The R Foundation (2020) R: the R project for statistical computing. https://www.r-project.org/. Accessed 11 May 2020
Trapnell C, Hendrickson DG, Sauvageau M et al (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31:46–53. https://doi.org/10.1038/nbt.2450
Uhe M, Hogekamp C, Hartmann RM et al (2018) The mycorrhiza-dependent defensin MtDefMd1 of Medicago truncatula acts during the late restructuring stages of arbusculecontaining cells. PLoS One 13:e0191841. https://doi.org/10.1371/journal.pone.0191841
Venkateshwaran M, Jayaraman D, Chabaud M et al (2015) A role for the mevalonate pathway in early plant symbiotic signaling. Proc Natl Acad Sci 112:9781–9786. https://doi.org/10.1073/pnas.1413762112
Vernié T, Camut S, Camps C et al (2016) PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through its ubiquitination activity in Medicago truncatula. Plant Physiol 170:2312–2324. https://doi.org/10.1104/pp.15.01694
Vierheilig H, Coughlan AP, Wyss U, Piché Y (1998) Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol 64:5004–5007. https://doi.org/10.1128/aem.64.12.5004-5007.1998
Vitousek PM, Porder S, Houlton BZ, Chadwick OA (2010) Terrestrial phosphorus limitation: mechanisms, implications, and nitrogen–phosphorus interactions. Ecol Appl 20:5–15. https://doi.org/10.1890/08-0127.1
Wang S, Chen A, Xie K et al (2020) Functional analysis of the OsNPF4.5 nitrate transporter reveals a conserved mycorrhizal pathway of nitrogen acquisition in plants. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.2000926117
Wang T, Zhao M, Zhang X et al (2017) Novel phosphate deficiency-responsive long non-coding RNAs in the legume model plant Medicago truncatula. J Exp Bot 68:5937–5948. https://doi.org/10.1093/jxb/erx384
Wang X, Zhao S, Bücking H (2016) Arbuscular mycorrhizal growth responses are fungal specific but do not differ between soybean genotypes with different phosphate efficiency. Ann Bot 118:11–21. https://doi.org/10.1093/aob/mcw074
Wipf D, Mongelard G, van Tuinen D et al (2014) Transcriptional responses of Medicago truncatula upon sulfur deficiency stress and arbuscular mycorrhizal symbiosis. Front Plant Sci 5:680. https://doi.org/10.3389/fpls.2014.00680
Wright DP, Read DJ, Scholes JD (1998) Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant, Cell Environ 21:881–891. https://doi.org/10.1046/j.1365-3040.1998.00351.x
Yoneyama K, Yoneyama K, Takeuchi Y, Sekimoto H (2007) Phosphorus deficiency in red clover promotes exudation of orobanchol, the signal for mycorrhizal symbionts and germination stimulant for root parasites. Planta 225:1031–1038. https://doi.org/10.1007/s00425-006-0410-1
Zhang Q, Blaylock LA, Harrison MJ (2010) Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell 22:1483–1497. https://doi.org/10.1105/tpc.110.074955
Zhang S, Daniels DA, Ivanov S et al (2022) A genetically encoded biosensor reveals spatiotemporal variation in cellular phosphate content in Brachypodium distachyon mycorrhizal roots. New Phytol. https://doi.org/10.1111/NPH.18081
Zhang YM, Wang HQ, Liu DM, Liu RJ (2020) Three tandemly aligned lea genes from Medicago truncatula confer differential protection to Escherichia coli against abiotic stresses. Biol Plant 64:95–103. https://doi.org/10.32615/bp.2019.112
Zheng C, Ji B, Zhang J et al (2015) Shading decreases plant carbon preferential allocation towards the most beneficial mycorrhizal mutualist. New Phytol 205:361–368. https://doi.org/10.1111/nph.13025
Acknowledgements
We would like to thank Lindsay McKeever for performing the Kjeldahl degradations for the 15N/14N analyses.
Funding
This work was funded by the USDA (2017–67014-26530), the SD Soybean Research and Promotion Council, and the Agricultural Experiment Station at SDSU to HB and SS. KG acknowledges the financial support of the North Carolina Agriculture Research Service (NCARS), the North Carolina Soybean Producers Association (2019–1656), and the USDA (2020–67013-31800).
Author information
Authors and Affiliations
Contributions
HB and SS conceived the original research plans; HB and AK designed the experiments; AK and JY performed the experiments, KRC and KG conducted the transcriptome analysis, and PP and GS completed the stable isotope analysis; KRC, AK, KG, JY, and PP analyzed the data; KRC wrote the article with input from all the authors; HB and SS supervised and completed the writing. HB agrees to serve as the author responsible for contact and ensures communication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cope, K.R., Kafle, A., Yakha, J.K. et al. Physiological and transcriptomic response of Medicago truncatula to colonization by high- or low-benefit arbuscular mycorrhizal fungi. Mycorrhiza 32, 281–303 (2022). https://doi.org/10.1007/s00572-022-01077-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00572-022-01077-2