Abstract
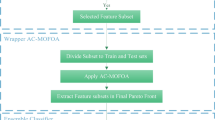
In microarray experiments, the sample size is considerably smaller than that of the feature size, thereby imposing the curse of dimensionality problem. To resolve this issue, evolutionary algorithms are often utilized. In this paper, a novel framework for feature selection and classification of the microarray data is presented. Initially, a statistical filter, namely ANOVA, is used to select the relevant genes (features) from the original set of genes. Then, an evolutionary wrapper-based approach utilizing the principles of enhanced Jaya (EJaya) algorithm and forest optimization algorithm (FOA) is proposed to find the optimal set of genes from the previously selected genes. The main objective of using EJaya is to tune the two important parameters, namely local seeding changes and global seeding changes of FOA. During the selection of the optimal set of genes, support vector machine is employed as a classifier to classify the microarray data. To perform a comprehensive experimental study, the proposed method is tested on both binary-class and multi-class microarray datasets. From the extensive result analysis, it has been observed that the proposed technique achieves better classification accuracy with considerably less number of features than that of the benchmark schemes.






Similar content being viewed by others

References
Algamal ZY, Lee MH (2018) A two-stage sparse logistic regression for optimal gene selection in high-dimensional microarray data classification. In: Advances in data analysis and classification. Springer, Berlin, pp 1–19
Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci 96(12):6745–6750
Alshamlan H, Badr G, Alohali Y (2015a) mRMR-ABC: a hybrid gene selection algorithm for cancer classification using microarray gene expression profiling. BioMed Res Int 2015:604910–604910
Alshamlan HM, Badr GH, Alohali YA (2015b) Genetic bee colony (GBC) algorithm: a new gene selection method for microarray cancer classification. Comput Biol Chem 56:49–60
Apolloni J, Leguizamón G, Alba E (2016) Two hybrid wrapper-filter feature selection algorithms applied to high-dimensional microarray experiments. Appl Soft Comput 38:922–932
Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ (2001) Mll translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 30(1):41
Baliarsingh SK, Vipsita S, Muhammad K, Dash B, Bakshi S (2019) Analysis of high-dimensional genomic data employing a novel bio-inspired algorithm. Appl Soft Comput 77:520–532
Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M et al (2001) Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci 98(24):13790–13795
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A (2015) Distributed feature selection: an application to microarray data classification. Appl Soft Comput 30:136–150
Chinnaswamy A, Srinivasan R (2016) Hybrid feature selection using correlation coefficient and particle swarm optimization on microarray gene expression data. In: Innovations in bio-inspired computing and applications. Springer, Cham, pp 229–239
Cho-Vega JH, Rassidakis GZ, Admirand JH, Oyarzo M, Ramalingam P, Paraguya A, McDonnell TJ, Amin HM, Medeiros LJ (2004) Mcl-1 expression in b-cell non-hodgkin’s lymphomas. Hum Pathol 35(9):1095–1100
Chouhan SS, Kaul A, Singh UP (2018a) Soft computing approaches for image segmentation: a survey. Multimed Tools Appl 77(21):28483–28537
Chouhan SS, Kaul A, Singh UP, Jain S (2018b) Bacterial foraging optimization based radial basis function neural network (BRBFNN) for identification and classification of plant leaf diseases: an automatic approach towards plant pathology. IEEE Access 6:8852–8863
Chuang JC, Stehr H, Liang Y, Das M, Huang J, Diehn M, Wakelee HA, Neal JW (2017) Erbb2-mutated metastatic non-small cell lung cancer: response and resistance to targeted therapies. J Thorac Oncol 12(5):833–842
Collins CT, Hess JL (2016) Role of hoxa9 in leukemia: dysregulation, cofactors and essential targets. Oncogene 35(9):1090
Dashtban M, Balafar M (2017) Gene selection for microarray cancer classification using a new evolutionary method employing artificial intelligence concepts. Genomics 109(2):91–107
Dwivedi AK (2018) Artificial neural network model for effective cancer classification using microarray gene expression data. Neural Comput Appl 29(12):1545–1554
Elyasigomari V, Lee D, Screen H, Shaheed M (2017) Development of a two-stage gene selection method that incorporates a novel hybrid approach using the cuckoo optimization algorithm and harmony search for cancer classification. J Biomed Inform 67:11–20
Eskinazi R, Thöny B, Svoboda M, Robberecht P, Dassesse D, Heizmann CW, Van Laethem JL, Resibois A (1999) Overexpression of pterin-4a-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 in human colon cancer. Am J Pathol 155(4):1105–1113
Ezejiofor IF, Adelusola K, Durosinmi MA, Leoncini L, Odesanmi WO, Ambrosio MR, Lazzi S, Olaofe RO, Gbutorano G et al (2018) Immunohistochemical characterization of small round blue cell tumors of childhood at ile-ife, Nigeria: a 10-year retrospective study. Arch Med Health Sci 6(1):64
Galani E, Sgouros J, Petropoulou C, Janinis J, Aravantinos G, Dionysiou-Asteriou D, Skarlos D, Gonos E (2002) Correlation of mdr-1, nm23-h1 and h sema e gene expression with histopathological findings and clinical outcome in ovarian and breast cancer patients. Anticancer Res 22(4):2275–2280
García-Nieto J, Alba E (2012a) Parallel multi-swarm optimizer for gene selection in DNA microarrays. Appl Intell 37(2):255–266
García-Nieto J, Alba E (2012b) Parallel multi-swarm optimizer for gene selection in DNA microarrays. Appl Intell 37(2):255–266
Ghaemi M, Feizi-Derakhshi MR (2014) Forest optimization algorithm. Exp Syst Appl 41(15):6676–6687
Ghosh M, Guha R, Sarkar R, Abraham A (2019) A wrapper-filter feature selection technique based on ant colony optimization. Neural Comput Appl. https://doi.org/10.1007/s00521-019-04171-3
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Hall MA (1999) Correlation-based feature selection for machine learning. Doctoral dissertation, The University of Waikato
Heit C, Jackson BC, McAndrews M, Wright MW, Thompson DC, Silverman GA, Nebert DW, Vasiliou V (2013) Update of the human and mouse serpin gene superfamily. Hum Genom 7(1):22
Hernandez JCH, Duval B, Hao JK (2007) A genetic embedded approach for gene selection and classification of microarray data. In: European conference on evolutionary computation, machine learning and data mining in bioinformatics, Springer, pp 90–101
Ibrahim AO, Shamsuddin SM, Abraham A, Qasem SN (2019) Adaptive memetic method of multi-objective genetic evolutionary algorithm for backpropagation neural network. Neural Comput Appl. https://doi.org/10.1007/s00521-018-03990-0
Jothi G, Inbarani HH, Azar AT, Devi KR (2018) Rough set theory with jaya optimization for acute lymphoblastic leukemia classification. Neural Comput Appl. https://doi.org/10.1007/s00521-018-3359-7
Jung JH, Jung CK, Choi HJ, Jun KH, Yoo J, Kang SJ, Lee KY (2009) Diagnostic utility of expression of claudins in non-small cell lung cancer: different expression profiles in squamous cell carcinomas and adenocarcinomas. Pathol Res Pract 205(6):409–416
Kar S, Sharma KD, Maitra M (2015) Gene selection from microarray gene expression data for classification of cancer subgroups employing PSO and adaptive K-nearest neighborhood technique. Exp Syst Appl 42(1):612–627
Kečo D, Subasi A, Kevric J (2018) Cloud computing-based parallel genetic algorithm for gene selection in cancer classification. Neural Comput Appl 30(5):1601–1610
Kim Y, Yoon S, Kim SJ, Kim JS, Cheong JW, Min YH (2012) Myeloperoxidase expression in acute myeloid leukemia helps identifying patients to benefit from transplant. Yonsei Med J 53(3):530–536
Lee CT, Chow NH, Su PF, Lin SC, Lin PC, Lee JC (2008) The prognostic significance of ron and met receptor coexpression in patients with colorectal cancer. Dis Colon Rectum 51(8):1268–1274
Li T, Zhang C, Ogihara M (2004) A comparative study of feature selection and multiclass classification methods for tissue classification based on gene expression. Bioinformatics 20(15):2429–2437
Liu KH, Zeng ZH, Ng VTY (2016) A hierarchical ensemble of ECOC for cancer classification based on multi-class microarray data. Inf Sci 349:102–118
Matthews BW (1975) Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim Biophys Acta (BBA) Protein Struct 405(2):442–451
Melhem R, Xx Zhu, Hailat N, Strahler JR, Hanash SM (1991) Characterization of the gene for a proliferation-related phosphoprotein (oncoprotein 18) expressed in high amounts in acute leukemia. J Biol Chem 266(27):17747–17753
Mohapatra P, Chakravarty S, Dash P (2016) Microarray medical data classification using kernel ridge regression and modified cat swarm optimization based gene selection system. Swarm Evolut Comput 28:144–160
Motieghader H, Najafi A, Sadeghi B, Masoudi-Nejad A (2017) A hybrid gene selection algorithm for microarray cancer classification using genetic algorithm and learning automata. Inform Med Unlocked 9:246–254
Mukhopadhyay A, Bandyopadhyay S, Maulik U (2010) Multi-class clustering of cancer subtypes through svm based ensemble of pareto-optimal solutions for gene marker identification. PloS One 5(11):e13803
Nash MA, Deavers MT, Freedman RS (2002) The expression of decorin in human ovarian tumors. Clin Cancer Res 8(6):1754–1760
Niu Q, Zhang H, Li K (2014a) An improved TLBO with elite strategy for parameters identification of PEM fuel cell and solar cell models. Int J Hydrog Energy 39(8):3837–3854
Niu Q, Zhang L, Li K (2014b) A biogeography-based optimization algorithm with mutation strategies for model parameter estimation of solar and fuel cells. Energy Convers Manag 86:1173–1185
Orujpour M, Feizi-Derakhshi MR, Rahkar-Farshi T (2019) Multi-modal forest optimization algorithm. Neural Comput Appl. https://doi.org/10.1007/s00521-019-04113-z
Pal NR, Aguan K, Sharma A, Amari Si (2007) Discovering biomarkers from gene expression data for predicting cancer subgroups using neural networks and relational fuzzy clustering. BMC Bioinform 8(1):5
Pang S, Havukkala I, Hu Y, Kasabov N (2007) Classification consistency analysis for bootstrapping gene selection. Neural Comput Appl 16(6):527–539
Petricoin EF III, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC et al (2002) Use of proteomic patterns in serum to identify ovarian cancer. Lancet 359(9306):572–577
Potharaju SP, Sreedevi M (2019) Distributed feature selection (DFS) strategy for microarray gene expression data to improve the classification performance. Clin Epidemiol Glob Health 7(2):171–176
Rao R (2016) Jaya: a simple and new optimization algorithm for solving constrained and unconstrained optimization problems. Int J Ind Eng Comput 7(1):19–34
Sharma A, Paliwal KK, Imoto S, Miyano S (2014) A feature selection method using improved regularized linear discriminant analysis. Mach Vis Appl 25(3):775–786
Sharma S, Kaul A (2018) Hybrid fuzzy multi-criteria decision making based multi cluster head dolphin swarm optimized IDS for VANET. Veh Commun 12:23–38
Sheskin DJ (2003) Handbook of parametric and nonparametric statistical procedures. CRC Press, Boca Raton
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45(4):427–437
Tabakhi S, Najafi A, Ranjbar R, Moradi P (2015) Gene selection for microarray data classification using a novel ant colony optimization. Neurocomputing 168:1024–1036
Tang B, Xiang K, Pang M (2018) An integrated particle swarm optimization approach hybridizing a new self-adaptive particle swarm optimization with a modified differential evolution. Neural Comput Appl. https://doi.org/10.1007/s00521-018-3878-2
Valdés-Mora F, Locke WJ, Bandrés E, Gallego-Ortega D, Cejas P, García-Cabezas MA, Colino-Sanguino Y, Feliú J, del Pulgar TG, Lacal JC (2017) Clinical relevance of the transcriptional signature regulated by cdc42 in colorectal cancer. Oncotarget 8(16):26755
Wang A, An N, Chen G, Yang J, Li L, Alterovitz G (2014a) Incremental wrapper based gene selection with Markov blanket. In: 2014 IEEE international conference on bioinformatics and biomedicine (BIBM). IEEE, pp 74–79
Wang X, Gotoh O (2009) Accurate molecular classification of cancer using simple rules. BMC Med Genom 2(1):64
Wang Y, Yang XG, Lu Y (2019) Informative gene selection for microarray classification via adaptive elastic net with conditional mutual information. Appl Math Model 71:286–297
Wang ZQ, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud MC, Sebastianelli A, Bachvarov D (2014b) Role of the polypeptide n-acetylgalactosaminyltransferase 3 in ovarian cancer progression: possible implications in abnormal mucin o-glycosylation. Oncotarget 5(2):544
Yagasaki F, Wakao D, Yokoyama Y, Uchida Y, Murohashi I, Kayano H, Taniwaki M, Matsuda A, Bessho M (2001) Fusion of etv6 to fibroblast growth factor receptor 3 in peripheral t-cell lymphoma with at (4; 12)(p16; p13) chromosomal translocation. Cancer Res 61(23):8371–8374
Yakirevich E, Resnick MB, Mangray S, Wheeler M, Jackson CL, Lombardo KA, Lee J, Kim KM, Gill AJ, Wang K et al (2016) Oncogenic alk fusion in rare and aggressive subtype of colorectal adenocarcinoma as a potential therapeutic target. Clin Cancer Res 22(15):3831–3840
Yu K, Wang X, Wang Z (2016) An improved teaching-learning-based optimization algorithm for numerical and engineering optimization problems. J Intell Manuf 27(4):831–843
Zhao H, Sun Q, Li L, Zhou J, Zhang C, Hu T, Zhou X, Zhang L, Wang B, Li B et al (2019) High expression levels of aggf1 and mfap4 predict primary platinum-based chemoresistance and are associated with adverse prognosis in patients with serous ovarian cancer. J Cancer 10(2):397
Zhao Y, Lu H, Yan A, Yang Y, Meng Q, Sun L, Pang H, Li C, Dong X, Cai L (2013) Abcc3 as a marker for multidrug resistance in non-small cell lung cancer. Sci Rep 3:3120
Zhu Z, Ong YS, Dash M (2007) Markov blanket-embedded genetic algorithm for gene selection. Pattern Recognit 40(11):3236–3248
Acknowledgements
This research is partially supported by the following Grant: Grant No. SR/FST/ETI-335/2013 by Fund for Improvement of S&T Infrastructure in Higher Educational Institutions (FIST) Program of Department of Science and Technology, Government of India to International Institute of Information Technology, Bhubaneswar, Odisha, India.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Baliarsingh, S.K., Vipsita, S. & Dash, B. A new optimal gene selection approach for cancer classification using enhanced Jaya-based forest optimization algorithm. Neural Comput & Applic 32, 8599–8616 (2020). https://doi.org/10.1007/s00521-019-04355-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-019-04355-x