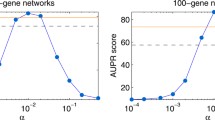
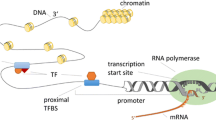
Abstract
The most basic molecular mechanism enabling a living cell to dynamically adapt to variation occurring in its intra and extracellular environment is constituted by its ability to regulate the expression of many of its genes. At biomolecular level, this ability is mainly due to interactions occurring between regulatory motifs located in the core promoter regions and the transcription factors. A crucial question investigated by recently published works is if, and at what extent, the transcription patterns of large sets of genes can be predicted using only information encoded in the promoter regions. Even if encouraging results were obtained in gene expression patterns prediction experiments the assumption that all the signals required for the regulation of gene expression are contained in the gene promoter regions is an oversimplification as pointed out by recent findings demonstrating the existence of many regulatory levels involved in the fine modulation of gene transcription levels. In this contribution, we investigate the potential improvement in gene expression prediction performances achievable by using early and late data integration methods in order to provide a complete overview of the capabilities of data fusion approaches in a problem that can be annoverated among the most difficult in modern bioinformatics.


Similar content being viewed by others
References
Beer M, Tavazoie S (2004) Predicting gene expression from sequence. Cell 117
desJardins M et al (1997) Prediction of enzyme classification from protein sequence without the use of sequence similarity. In: Proceedings of the 5th international conference on intelligent systems for molecular biology. AAAI Press, Menlo Park, pp 92–99
Friedman J et al (2000) Additive logistic regression: a statistical view of boosting. Ann Stat 38(2):337–374
Gasch P et al (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11:4241–4257
Hartigan J (1975) Clustering algorithms. Wiley, New York
Iorio F et al (2009) Identifying network of drug mode action by gene expression profiling. J Comput Biol 16
Kuncheva LI et al (2001) Decision templates for multiple classifier fusion: an experimental comparison. Pattern Recognit 34(2):299–314
Lamb J et al (2006) The connectivity map: using gene-expression signatures to connect small molecules genes and diseases. Science 313
Lanckriet G et al (2004) A statistical framework for genomic data fusion. Bioinformatics 20:2626–2635
Lin H, Lin C, Weng R (2007) A note on Platt’s probabilistic outputs for support vector machines. Mach Learn 68:267–276
McIsaac K et al (2006) An improved map of conserved regulatory sites map for Saccharomyces cerevisiae. BMC Bioinf 7
Millar C, Grunstein M (2006) Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol 7
Noble W, Ben-Hur A (2007) Integrating information for protein function prediction. In: Lengauer T (ed) m genomes to therapies, vol 3, Wiley, New York, pp 1297–1314
O’Connor T, Wryck J (2007) Chromatindb: a database of genome-wide histone modification patterns for saccharomyces cerevisiae. Bioinformatics 23
Pavesi G, Valentini G (2009) Classification of co-expressed genes from dna regulatory regions. Information Fusion 10
Pavlidis P et al (2002) Learning gene functional classification from multiple data. J Comput Biol 9
Rosset S et al (2004) Boosting as a regularized path to a maximum margin classifier. J Mach Learn Res 5
Spellman P et al (1998) Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomices cerevisiae by microarray hybridization. Mol Biol Cell 9:3273–3297
Subramanian A et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102
Yuan Y et al (2007) Prediction gene expression from sequence: a reexamination. PLOS Comp Biol 3
Zhu J et al (2004) Multi-class adaboost. Statistics and its Interface 2
Acknowledgments
The authors would like to gratefully acknowledge partial support by the PASCAL2 Network of Excellence under EC grant no. 216886. This publication only reflects the authors’ views. The author would also like to expressly thank Giorgio Valentini for the examination of early versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Re, M. Comparing early and late data fusion methods for gene expression prediction. Soft Comput 15, 1497–1504 (2011). https://doi.org/10.1007/s00500-010-0599-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00500-010-0599-6