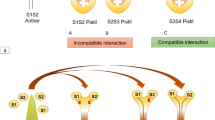
Key message
Epigenetic dominance modifier.
Abstract
In polymorphic loci, complex genetic dominance relationships between alleles are often observed. In plants, control of self-incompatibility (SI) expression via allelic interactions in the Brassicaceae is the best-known example of such mechanisms. Here, with emphasis on two recently published papers, we review the progress toward understanding the dominance regulatory mechanism of SI in the Brassicaceae. Multiple small RNA genes linked to the Self-incompatibility (S) locus were found in both Brassica and Arabidopsis genera. Mono-allelic gene expression of the male determinant of SI, SP11/SCR, from a dominant S-allele is under epigenetic control by such small RNA genes. Possible evolutionary trajectories leading to the formation of multilayered dominance hierarchy in Brassicaceae are discussed. We also identify some remaining questions for future studies.

Similar content being viewed by others
References
Alleman M, Sidorenko L, McGinnis K et al (2006) An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442:295–298. https://doi.org/10.1038/nature04884
Allen E, Xie Z, Gustafson AM et al (2004) Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet 36:1282–1290. https://doi.org/10.1038/ng1478
Billiard S, Castric V (2011) Evidence for Fisher’s dominance theory: how many “special cases”? Trends Genet 27:441–445. https://doi.org/10.1016/j.tig.2011.06.005
Brennan AC, Harris SA, Tabah DA, Hiscock SJ (2002) The population genetics of sporophytic self-incompatibility in Senecio squalidus L. (Asteraceae) I: S allele diversity in a natural population. Heredity (Edinb) 89:430–438. https://doi.org/10.1038/sj.hdy.6800159
Castric V, Vekemans X (2007) Evolution under strong balancing selection: how many codons determine specificity at the female self-incompatibility gene SRK in Brassicaceae? BMC Evol Biol 7:132. https://doi.org/10.1186/1471-2148-7-132
Durand E, Meheust R, Soucaze M et al (2014) Dominance hierarchy arising from the evolution of a complex small RNA regulatory network. Science (80-) 346:1200–1205. https://doi.org/10.1126/science.1259442
Edlund AF, Swanson R, Preuss D (2004) Pollen and stigma structure and function: the role of diversity in pollination. Plant Cell 16(Suppl):S84–S97. https://doi.org/10.1105/tpc.015800
Erhard KF, Stonaker JL, Parkinson SE et al (2009) RNA polymerase iv functions in paramutation in Zea mays. Science (80-) 323:1201–1205. https://doi.org/10.1126/science.1164508
Finnegan EJ, Liang D, Wang MB (2011) Self-incompatibility: Smi silences through a novel sRNA pathway. Trends Plant Sci 16:238–241. https://doi.org/10.1016/j.tplants.2011.01.002
Fujii S, Kubo K, Takayama S (2016) Non-self- and self-recognition models in plant self-incompatibility. Nat Plants 2:16130. https://doi.org/10.1038/nplants.2016.130
Goubet PM, Bergès H, Bellec A et al (2012) Contrasted patterns of molecular evolution in dominant and recessive self-incompatibility haplotypes in Arabidopsis. PLoS Genet 8:e1002495. https://doi.org/10.1371/journal.pgen.1002495
Hatakeyama K, Watanabe M, Takasaki T et al (1998) Dominance relationships between S-alleles in self-incompatible Brassica campestris L. Heredity (Edinb) 80:241–247
Igic B, Lande R, Kohn JR (2008) Loss of self-incompatibility and its evolutionary consequences. Int J Plant Sci 169:93–104. https://doi.org/10.1086/523362
Iwano M, Takayama S (2012) Self/non-self discrimination in angiosperm self-incompatibility. Curr Opin Plant Biol 15:78–83. https://doi.org/10.1016/j.pbi.2011.09.003
Iwano M, Shiba H, Funato M et al (2003) Immunohistochemical studies on translocation of pollen S-haplotype determinant in self-incompatibility of Brassica rapa. Plant Cell Physiol 44:428–436. https://doi.org/10.1093/pcp/pcg056
Kakizaki T, Takada Y, Ito A et al (2003) Linear dominance relationship among four class-II S haplotypes in pollen is determined by the expression of SP11 in Brassica self-incompatibility. Plant Cell Physiol 44:70–75. https://doi.org/10.1093/pcp/pcg009
Kinoshita T, Miura A, Choi Y et al (2004) One-way control of FWA imprinting in Arabidopsis endosperm by DNA methylation. Science 303:521–523. https://doi.org/10.1126/science.1089835
Kowyama Y, Takahasi H, Muraoka K et al (1994) Number, frequency and dominance relationships of S-alleles in diploid Ipomoea trifida. Heredity (Edinb) 73:275–283. https://doi.org/10.1038/hdy.1994.134
Liu L, Fan X (2013) Tapetum: regulation and role in sporopollenin biosynthesis in Arabidopsis. Plant Mol Biol 83:165–175. https://doi.org/10.1007/s11103-013-0085-5
Llaurens V, Billiard S, Leducq J-B et al (2008) Does frequency-dependent selection with complex dominance interactions accurately predict allelic frequencies at the self-incompatibility locus in Arabidopsis halleri? Evolution 62:2545–2557. https://doi.org/10.1111/j.1558-5646.2008.00469.x
Mable BK, Schierup MH, Charlesworth D (2003) Estimating the number, frequency, and dominance of S-alleles in a natural population of Arabidopsis lyrata (Brassicaceae) with sporophytic control of self-incompatibility. Heredity (Edinb) 90:422–431. https://doi.org/10.1038/sj.hdy.6800261
Nasrallah JB, Nasrallah ME (1993) Pollen[mdash]Stigma signaling in the sporophytic self-incompatibility response. Plant Cell 5:1325–1335. https://doi.org/10.1105/tpc.5.10.1325
Prigoda NL, Nassuth A, Mable BK (2005) Phenotypic and genotypic expression of self-incompatibility haplotypes in Arabidopsis lyrata suggests unique origin of alleles in different dominance classes. Mol Biol Evol 22:1609–1620. https://doi.org/10.1093/molbev/msi153
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425. https://doi.org/10.1101/gad.1476406
Schopfer CR, Nasrallah ME, Nasrallah JB (1999) The male determinant of self-incompatibility in Brassica. Science 286:1697–1700. https://doi.org/10.1126/science.286.5445.1697
Scott RJ, Spielman M, Dickinson HG (2004) Stamen structure and function. Plant Cell 16(Suppl):S46–S60. https://doi.org/10.1105/tpc.017012
Shabalina SA, Koonin EV (2008) Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol 23:578–587. https://doi.org/10.1016/j.tree.2008.06.005
Shiba H, Takayama S, Iwano M et al (2001) A pollen coat protein, SP11/SCR, determines the pollen S-specificity in the self-incompatibility of Brassica species. Plant Physiol 125:2095–2103
Shiba H, Iwano M, Entani T et al (2002) The dominance of alleles controlling self-incompatibility in Brassica pollen is regulated at the RNA level. Plant Cell 14:491–504. https://doi.org/10.1105/tpc.010378
Shiba H, Kakizaki T, Iwano M et al (2006) Dominance relationships between self-incompatibility alleles controlled by DNA methylation. Nat Genet 38:297–299. https://doi.org/10.1038/ng1734
Suzuki G, Kai N, Hirose T et al (1999) Genomic organization of the S locus: Identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics 153:391–400
Takayama S, Isogai A (2005) Self-incompatibility in plants. Annu Rev Plant Biol 56:467–489. https://doi.org/10.1146/annurev.arplant.56.032604.144249
Takayama S, Shiba H, Iwano M et al (2000) The pollen determinant of self-incompatibility in Brassica campestris. Proc Natl Acad Sci USA 97:1920–1925. https://doi.org/10.1073/pnas.040556397
Takayama S, Shimosato H, Shiba H et al (2001) Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413:534–538. https://doi.org/10.1038/35097104
Tarutani Y, Shiba H, Iwano M et al (2010) Trans -acting small RNA determines dominance relationships in Brassica self-incompatibility. Nature 466:983–986. https://doi.org/10.1038/nature09308
Thompson KF, Taylor JP (1966) Non-linear dominance relationships between S alleles. Heredity (Edinb) 21:345–362
Visser DL, Van Hal JG, Verhoeven WH (1982) Classification of S-alleles by their activity in S-heterozygotes of brussels sprouts (Brassica oleracea var. Gemmifera (DC.) S chultz). Euphytica 31:603–611. https://doi.org/10.1007/BF00039198
Yasuda S, Wada Y, Kakizaki T et al (2016) A complex dominance hierarchy is controlled by polymorphism of small RNAs and their targets. Nat Plants 3:16206. https://doi.org/10.1038/nplants.2016.206
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas (23113002, 16H06467, 16H06464 to S.T.; 16H01467 to S.F.), Grants-in-Aid for Scientific Research (21248014, 25252021, 16H06380 to S.T.), Grant-in-Aid for Challenging Exploratory Research (15K14626 to S.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), and Japan Science and Technology Agency (JST) PRESTO program (JPMJPR16Q8) to S.F.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Mengxiang Sun.
A contribution to the special issue ‘Plant Reproduction Research in Asia’.
Rights and permissions
About this article
Cite this article
Fujii, S., Takayama, S. Multilayered dominance hierarchy in plant self-incompatibility. Plant Reprod 31, 15–19 (2018). https://doi.org/10.1007/s00497-017-0319-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00497-017-0319-9