Abstract
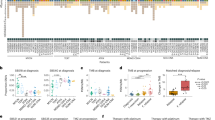
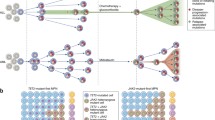
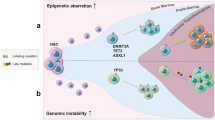
The notion of cancer as a complex evolutionary system has been validated by in-depth molecular analyses of tumor progression over the last years. While a complex interplay of cell-autonomous programs and cell-cell interactions determines proliferation and differentiation during normal development, intrinsic and acquired plasticity of cancer cells allow for evasion of growth factor limitations, apoptotic signals, or attacks from the immune system. Treatment-induced molecular selection processes have been described by a number of studies already, but understanding of those events facilitating metastatic spread, organ-specific homing, and resistance to anoikis is still in its early days. In principle, somatic events giving rise to cancer progression should be easier to follow in childhood tumors bearing fewer mutations and genomic aberrations than their counterparts in adulthood. We have previously reported on the genetic events accompanying relapsing neuroblastoma, a solid tumor of early childhood. Our results indicated significantly higher single nucleotide variants in relapse tumors, gave hints for branched tumor evolution upon treatment and clonal selection as deduced from shifts in allelic frequencies between primary and relapsing neuroblastoma. Here, we will review these findings and give an outlook on dealing with intratumoral heterogeneity and sub-clonal diversity in neuroblastoma for future targeted treatments.



Similar content being viewed by others
References
Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, Le Quesne J, Moore DA, Veeriah S, Rosenthal R, Marafioti T, Kirkizlar E, Watkins TBK, McGranahan N, Ward S, Martinson L, Riley J, Fraioli F, Al Bakir M, Grönroos E, Zambrana F, Endozo R, Bi WL, Fennessy FM, Sponer N, Johnson D, Laycock J, Shafi S, Czyzewska-Khan J, Rowan A, Chambers T, Matthews N, Turajlic S, Hiley C, Lee SM, Forster MD, Ahmad T, Falzon M, Borg E, Lawrence D, Hayward M, Kolvekar S, Panagiotopoulos N, Janes SM, Thakrar R, Ahmed A, Blackhall F, Summers Y, Hafez D, Naik A, Ganguly A, Kareht S, Shah R, Joseph L, Marie Quinn A, Crosbie PA, Naidu B, Middleton G, Langman G, Trotter S, Nicolson M, Remmen H, Kerr K, Chetty M, Gomersall L, Fennell DA, Nakas A, Rathinam S, Anand G, Khan S, Russell P, Ezhil V, Ismail B, Irvin-Sellers M, Prakash V, Lester JF, Kornaszewska M, Attanoos R, Adams H, Davies H, Oukrif D, Akarca AU, Hartley JA, Lowe HL, Lock S, Iles N, Bell H, Ngai Y, Elgar G, Szallasi Z, Schwarz RF, Herrero J, Stewart A, Quezada SA, Peggs KS, Van Loo P, Dive C, Lin CJ, Rabinowitz M, Aerts HJWL, Hackshaw A, Shaw JA, Zimmermann BG, TRACERx consortium; PEACE consortium, Swanton C (2017) Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545:446
Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jager N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, Lopez-Otin C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdes-Mas R, van Buuren MM, van’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR (2013) Signatures of mutational processes in human cancer. Nature 500:415–421
Althoff K, Beckers A, Bell E, Nortmeyer M, Thor T, Sprussel A, Lindner S, De Preter K, Florin A, Heukamp LC, Klein-Hitpass L, Astrahantseff K, Kumps C, Speleman F, Eggert A, Westermann F, Schramm A, Schulte JH (2015) A Cre-conditional MYCN-driven neuroblastoma mouse model as an improved tool for preclinical studies. Oncogene 34:3357–3368
Bardelli A, Cahill DP, Lederer G, Speicher MR, Kinzler KW, Vogelstein B, Lengauer C (2001) Carcinogen-specific induction of genetic instability. Proc Natl Acad Sci U S A 98:5770–5775
Bellini A, Bernard V, Leroy Q, Rio Frio T, Pierron G, Combaret V, Lapouble E, Clement N, Rubie H, Thebaud E, Chastagner P, Defachelles AS, Bergeron C, Buchbinder N, Taque S, Auvrignon A, Valteau-Couanet D, Michon J, Janoueix-Lerosey I, Delattre O, Schleiermacher G (2015) Deep sequencing reveals occurrence of subclonal ALK mutations in neuroblastoma at diagnosis. Clin Cancer Res 21:4913–4921
Cahill DP, Kinzler KW, Vogelstein B, Lengauer C (1999) Genetic instability and Darwinian selection in tumours. Trends Cell Biol 9:M57–M60
Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR (2008) Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci U S A 105:13081–13086
Chicard M, Boyault S, Colmet Daage L, Richer W, Gentien D, Pierron G, Lapouble E, Bellini A, Clement N, Iacono I, Bréjon S, Carrere M, Reyes C, Hocking T, Bernard V, Peuchmaur M, Corradini N, Faure-Conter C, Coze C, Plantaz D, Defachelles AS, Thebaud E, Gambart M, Millot F, Valteau-Couanet D, Michon J, Puisieux A, Delattre O, Combaret V, Schleiermacher G (2016) Genomic copy number profiling using circulating free tumor DNA highlights heterogeneity in neuroblastoma. Clin Cancer Res 22(22):5564–5573. https://doi.org/10.1158/1078-0432.CCR-16-0500
Chicard M, Colmet Daage L, Clement N, Danzon A, Bohec M, Bernard V, Baulande S, Bellini A, Deveau P, Pierron G, Lapouble E, Janoueix-Lerosey I, Peuchmaur M, Corradini N, Desfachelles AS, Valteau-Couanet D, Michon J, Combaret V, Delattre O, Schleiermacher G (2017) Whole exome sequencing of cell-free DNA reveals temporo-spatial heterogeneity and identifies treatment-resistant clones in neuroblastoma. Clinical Cancer Res. https://doi.org/10.1158/1078-0432.CCR-17-1586
Eleveld TF, Oldridge DA, Bernard V, Koster J, Daage LC, Diskin SJ, Schild L, Bentahar NB, Bellini A, Chicard M, Lapouble E, Combaret V, Legoix-Ne P, Michon J, Pugh TJ, Hart LS, Rader J, Attiyeh EF, Wei JS, Zhang S, Naranjo A, Gastier-Foster JM, Hogarty MD, Asgharzadeh S, Smith MA, Guidry Auvil JM, Watkins TBK, Zwijnenburg DA, Ebus ME, van Sluis P, Hakkert A, van Wezel E, van der Schoot CE, Westerhout EM, Schulte JH, Tytgat GA, Dolman MEM, Janoueix-Lerosey I, Gerhard DS, Caron HN, Delattre O, Khan J, Versteeg R, Schleiermacher G, Molenaar JJ, Maris JM (2015) Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet 47:864–871
Gao Y, Ferguson DO, Xie W, Manis JP, Sekiguchi J, Frank KM, Chaudhuri J, Horner J, DePinho RA, Alt FW (2000) Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897
Greaves M, Maley CC (2012) Clonal evolution in cancer. Nature 481:306–313
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674
Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, Valteau-Couanet D, Frebourg T, Michon J, Lyonnet S, Amiel J, Delattre O (2008) Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455:967–970
Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, Kinzler KW, Vogelstein B, Willis J, Markowitz SD (2008) Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A 105:4283–4288
Loeb LA (2011) Human cancers express mutator phenotypes: origin, consequences and targeting. Nat Rev Cancer 11:450–457
Ma X, Edmonson M, Yergeau D, Muzny DM, Hampton OA, Rusch M, Song G, Easton J, Harvey RC, Wheeler DA, Ma J, Doddapaneni H, Vadodaria B, Wu G, Nagahawatte P, Carroll WL, Chen IM, Gastier-Foster JM, Relling MV, Smith MA, Devidas M, Guidry Auvil JM, Downing JR, Loh ML, Willman CL, Gerhard DS, Mullighan CG, Hunger SP, Zhang J (2015) Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun 6:6604
Malinowska-Ozdowy K, Frech C, Schonegger A, Eckert C, Cazzaniga G, Stanulla M, zur Stadt U, Mecklenbrauker A, Schuster M, Kneidinger D, von Stackelberg A, Locatelli F, Schrappe M, Horstmann MA, Attarbaschi A, Bock C, Mann G, Haas OA, Panzer-Grumayer R (2015) KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia 29:1656–1667
McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, Birkbak NJ, Veeriah S, Van Loo P, Herrero J, Swanton C (2017) Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. https://doi.org/10.1016/j.cell.2017.10.001
Molenaar JJ, Koster J, Ebus ME, van Sluis P, Westerhout EM, de Preter K, Gisselsson D, Øra I, Speleman F, Caron HN, Versteeg R (2012) Copy number defects of G1-cell cycle genes in neuroblastoma are frequent and correlate with high expression of E2F target genes and a poor prognosis. Genes Chromosom Cancer 51:10–19
Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, Laureys G, Speleman F, Kim C, Hou C, Hakonarson H, Torkamani A, Schork NJ, Brodeur GM, Tonini GP, Rappaport E, Devoto M, Maris JM (2008) Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455:930–935
Nowell PC (1976) The clonal evolution of tumor cell populations. Science (New York, NY) 194:23–28
Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, Kim J, Lawrence MS, Lichenstein L, McKenna A, Pedamallu CS, Ramos AH, Shefler E, Sivachenko A, Sougnez C, Stewart C, Ally A, Birol I, Chiu R, Corbett RD, Hirst M, Jackman SD, Kamoh B, Khodabakshi AH, Krzywinski M, Lo A, Moore RA, Mungall KL, Qian J, Tam A, Thiessen N, Zhao Y, Cole KA, Diamond M, Diskin SJ, Mosse YP, Wood AC, Ji L, Sposto R, Badgett T, London WB, Moyer Y, Gastier-Foster JM, Smith MA, Guidry Auvil JM, Gerhard DS, Hogarty MD, Jones SJ, Lander ES, Gabriel SB, Getz G, Seeger RC, Khan J, Marra MA, Meyerson M, Maris JM (2013) The genetic landscape of high-risk neuroblastoma. Nat Genet 45:279–284
Sausen M, Leary RJ, Jones S, Wu J, Reynolds CP, Liu X, Blackford A, Parmigiani G, Diaz LA, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE, Hogarty MD (2013) Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet 45:12–17
Schilling FH, Spix C, Berthold F, Erttmann R, Fehse N, Hero B, Klein G, Sander J, Schwarz K, Treuner J, Zorn U, Michaelis J (2002) Neuroblastoma screening at one year of age. N Engl J Med 346:1047–1053
Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, Klijanienko J, Couturier J, Pierron G, Mosseri V, Valent A, Auger N, Plantaz D, Rubie H, Valteau-Couanet D, Bourdeaut F, Combaret V, Bergeron C, Michon J, Delattre O (2010) Accumulation of segmental alterations determines progression in neuroblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 28:3122–3130
Schleiermacher G, Javanmardi N, Bernard V, Leroy Q, Cappo J, Rio Frio T, Pierron G, Lapouble E, Combaret V, Speleman F, de Wilde B, Djos A, Ora I, Hedborg F, Trager C, Holmqvist BM, Abrahamsson J, Peuchmaur M, Michon J, Janoueix-Lerosey I, Kogner P, Delattre O, Martinsson T (2014) Emergence of new ALK mutations at relapse of neuroblastoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 32:2727–2734
Schramm A, Koster J, Assenov Y, Althoff K, Peifer M, Mahlow E, Odersky A, Beisser D, Ernst C, Henssen AG, Stephan H, Schroder C, Heukamp L, Engesser A, Kahlert Y, Theissen J, Hero B, Roels F, Altmuller J, Nurnberg P, Astrahantseff K, Gloeckner C, De Preter K, Plass C, Lee S, Lode HN, Henrich KO, Gartlgruber M, Speleman F, Schmezer P, Westermann F, Rahmann S, Fischer M, Eggert A, Schulte JH (2015) Mutational dynamics between primary and relapse neuroblastomas. Nat Genet 47:872–877
Schwartz M, Zlotorynski E, Kerem B (2006) The molecular basis of common and rare fragile sites. Cancer Lett 232:13–26
Siegmund KD, Marjoram P, Woo YJ, Tavare S, Shibata D (2009) Inferring clonal expansion and cancer stem cell dynamics from DNA methylation patterns in colorectal cancers. Proc Natl Acad Sci U S A 106:4828–4833
Stratton MR (2011) Exploring the genomes of cancer cells: progress and promise. Science (New York, NY) 331:1553–1558
Strauss R, Hamerlik P, Lieber A, Bartek J (2012) Regulation of stem cell plasticity: mechanisms and relevance to tissue biology and cancer. Mol Ther 20:887–897
Tweddle DA, Pearson ADJ, Haber M, Norris MD, Xue C, Flemming C, Lunec J (2003) The p53 pathway and its inactivation in neuroblastoma. Cancer Lett 197:93–98
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA (2015) Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (New York, NY) 350:207–211
van Groningen T, Koster J, Valentijn LJ, Zwijnenburg DA, Akogul N, Hasselt NE, Broekmans M, Haneveld F, Nowakowska NE, Bras J, van Noesel CJM, Jongejan A, van Kampen AH, Koster L, Baas F, van Dijk-Kerkhoven L, Huizer-Smit M, Lecca MC, Chan A, Lakeman A, Molenaar P, Volckmann R, Westerhout EM, Hamdi M, van Sluis PG, Ebus ME, Molenaar JJ, Tytgat GA, Westerman BA, van Nes J, Versteeg R (2017) Neuroblastoma is composed of two super-enhancer-associated differentiation states. Nat Genet 49:1261–1266
Van Roy N, Van Der Linden M, Menten B, Dheedene A, Vandeputte C, Van Dorpe J, Laureys G, Renard M, Sante T, Lammens T, De Wilde B, Speleman F, De Preter K (2017) Shallow whole genome sequencing on circulating cell-free DNA allows reliable noninvasive copy-number profiling in neuroblastoma patients. Clinical cancer research : an official journal of the American Association for Cancer Research 23:6305–6314
Versteeg R, van der Minne C, Plomp A, Sijts A, van Leeuwen A, Schrier P (1990) N-myc expression switched off and class I human leukocyte antigen expression switched on after somatic cell fusion of neuroblastoma cells. Mol Cell Biol 10:5416–5423
Yoshimura M, Itasaka S, Harada H, Hiraoka M (2013) Microenvironment and radiation therapy. Biomed Res Int 2013:685308
Acknowledgments
Work of AS and SR is supported by the DFG within Collaborative Research Center SFB876, TP C1 and the BMBF as part of the program “SYSMED-NB”, TP A2 and B3.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Schulte, M., Köster, J., Rahmann, S. et al. Cancer evolution, mutations, and clonal selection in relapse neuroblastoma. Cell Tissue Res 372, 263–268 (2018). https://doi.org/10.1007/s00441-018-2810-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00441-018-2810-5