Abstract
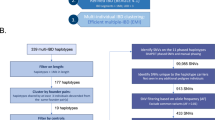
Familial adult myoclonus epilepsy (FAME) is a rare autosomal dominant disorder characterized by adult onset, involuntary muscle jerks, cortical myoclonus and occasional seizures. FAME is genetically heterogeneous with more than 70 families reported worldwide and five potential disease loci. The efforts to identify potential causal variants have been unsuccessful in all but three families. To date, linkage analysis has been the main approach to find and narrow FAME critical regions. We propose an alternative method, pedigree free identity-by-descent (IBD) mapping, that infers regions of the genome between individuals that have been inherited from a common ancestor. IBD mapping provides an alternative to linkage analysis in the presence of allelic and locus heterogeneity by detecting clusters of individuals who share a common allele. Succeeding IBD mapping, gene prioritization based on gene co-expression analysis can be used to identify the most promising candidate genes. We performed an IBD analysis using high-density single nucleotide polymorphism (SNP) array data followed by gene prioritization on a FAME cohort of ten European families and one Australian/New Zealander family; eight of which had known disease loci. By identifying IBD regions common to multiple families, we were able to narrow the FAME2 locus to a 9.78 megabase interval within 2p11.2–q11.2. We provide additional evidence of a founder effect in four Italian families and allelic heterogeneity with at least four distinct founders responsible for FAME at the FAME2 locus. In addition, we suggest candidate disease genes using gene prioritization based on gene co-expression analysis.




Similar content being viewed by others
References
Aerts S, Lambrechts D, Maity S et al (2006) Gene prioritization through genomic data fusion. Nat Biotechnol 24:537–544. doi:10.1038/nbt1203
Albrechtsen A, Sand Korneliussen T, Moltke I, van Overseem Hansen T, Nielsen FC, Nielsen R (2009) Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium. Genet Epidemiol 33:266–274. doi:10.1002/gepi.20378
Anvar SY, Khachatryan L, Vermaat M et al (2014) Determining the quality and complexity of next-generation sequencing data without a reference genome. Genome Biol 15:555. doi:10.1186/s13059-014-0555-3
Brin S, Page L (1998) The anatomy of a large-scale hypertextual web search engine. Comput Netw ISDN 30:107–117. doi:10.1016/S0169-7552(98)00110-X
Brown MD, Glazner CG, Zheng C, Thompson EA (2012) Inferring coancestry in population samples in the presence of linkage disequilibrium. Genetics 190(4):1447–1460
Browning SR, Browning BL (2010) High-resolution detection of identity by descent in unrelated individuals. Am J Hum Genet 86:526–539. doi:10.1016/j.ajhg.2010.02.021
Choo KHA (1998) Why is the centromere so cold? Genome Res 8:81–82
Crompton DE, Sadleir LG, Bromhead CJ et al (2012) Familial adult myoclonic epilepsy: recognition of mild phenotypes and refinement of the 2q locus. Arch Neurol-Chicago 69:474–481. doi:10.1001/archneurol.2011.584
De Fusco M, Vago R, Striano P et al (2014) The α2B-adrenergic receptor is mutant in cortical myoclonus and epilepsy. Ann Neurol 75:77–87. doi:10.1002/ana.24028
Depienne C, Magnin E, Bouteiller D et al (2010) Familial cortical myoclonic tremor with epilepsy: the third locus (FCMTE3) maps to 5p. Neurology 74:2000–2003. doi:10.1212/WNL.0b013e3181e396a8
Elia M, Musumeci SA, Ferri R et al (1998) Familial cortical tremor, epilepsy, and mental retardation: a distinct clinical entity? Arch Neurol 55:1569–1573
Freytag S, Gagnon-Bartsch J, Speed TP, Bahlo M (2015) Systematic noise degrades gene co-expression signals but can be corrected. BMC Bioinf 16:309. doi:10.1186/s12859-015-0745-3
Guerrini R, Bonanni P, Patrignani A et al (2001) Autosomal dominant cortical myoclonus and epilepsy (ADCME) with complex partial and generalized seizures: a newly recognized epilepsy syndrome with linkage to chromosome 2p11.1-q12.2. Brain 124:2459–2475
Henden L, Wakeham D, Bahlo M (2016) XIBD: software for inferring pairwise identity by descent on the X chromosome. Bioinformatics. doi:10.1093/bioinformatics/btw124
Ikeda A, Kakigi R, Funai N et al (1990) Cortical tremor: a variant of cortical reflex myoclonus. Neurology 40(10):1561–1565
Kang HJ, Kawasawa YI, Cheng F et al (2011) Spatio-temporal transcriptome of the human brain. Nature 478:483–489. doi:10.1038/nature10523
Koboldt DC, Steinberg KM, Larson DE, Wilson RK, Mardis ER (2013) The next-generation sequencing revolution and its impact on genomics. Cell 155:27–38. doi:10.1016/j.cell.2013.09.006
Licchetta L, Pippucci T, Bisulli F et al (2013) A novel pedigree with familial cortical myoclonic tremor and epilepsy (FCMTE): clinical characterization, refinement of the FCMTE2 locus, and confirmation of a founder haplotype. Epilepsia 54:298–1306. doi:10.1111/epi.12216
Madia F, Striano P, Di Bonaventura C et al (2008) Benign adult familial myoclonic epilepsy (BAFME): evidence of an extended founder haplotype on chromosome 2p11.1-q12.2 in five Italian families. Neurogenetics 9:139–142. doi:10.1007/s10048-008-0118-4
Mikami M, Yasuda T, Terao A et al (1999) Localization of a gene for benign adult familial myoclonic epilepsy to chromosome 8q23.3-q24.1. Am J Hum Genet 65:745–751
Mori S, Nakamura M, Yasuda T, Ueno SI, Kaneko S, Sano A (2011) Remapping and mutation analysis of benign adult familial myoclonic epilepsy in a Japanese pedigree. J Hum Genet 56:742–747. doi:10.1038/jhg.2011.93
Nava C, Keren B, Mignot C et al (2014) Prospective diagnostic analysis of copy number variants using SNP microarrays in individuals with autism spectrum disorders. Euro J Hum Genet 22:71–78. doi:10.1038/ejhg.2013.88
Niesler B, Frank B, Kapeller J, Rappold GA (2003) Cloning, physical mapping and expression analysis of the human 5-HT3 serotonin receptor-like genes HTR3C, HTR3D and HTR3E. Gene 310:101–111. doi:10.1016/S0378-1119(03)00503-1
Oliver KL, Lukic V, Thorne NP, Berkovic SF, Scheffer IE, Bahlo M (2014) Harnessing gene expression networks to prioritize candidate epileptic encephalopathy genes. PLoS One 9:e102079. doi:10.1371/journal.pone.0102079
Plaster NM, Uyama E, Uchino M, Ikeda T, Flanigan KM, Kondo I, Ptácek LJ (1999) Genetic localization of the familial adult myoclonic epilepsy (FAME) gene to chromosome 8q24. Neurology 53:1180–1183
Rabiner LR (1989) A tutorial on hidden Markov models and selected applications in speech recognition. IEEE 77:257–286
Saint-Martin C, Bouteiller D, Stevanin G et al (2008) Refinement of the 2p11.1-q12.2 locus responsible for cortical tremor associated with epilepsy and exclusion of candidate genes. Neurogenetics 9:69–71. doi:10.1007/s10048-007-0107-z
Stogmann E, Reinthaler E, Eltawil S et al (2013) Autosomal recessive cortical myoclonic tremor and epilepsy: association with a mutation in the potassium channel associated gene CNTN2. Brain 136:1155–1160. doi:10.1093/brain/awt068
Striano P, Madia F, Minetti C, Striano S, Zara F (2005) Electroclinical and genetic findings in a family with cortical tremor, myoclonus, and epilepsy. Epilepsia 46:1993–1995. doi:10.1111/j.1528-1167.2005.00346.x
The International HapMap 3 Consortium (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58
Uebele VN, Lagrutta A, Wade T et al (2000) Cloning and functional expression of two families of β-subunits of the large conductance calcium-activated K+ channel. J Biol Chem 375:23211–23218. doi:10.1074/jbc.M910187199
Wada Y, Shiraishi J, Nakamura M, Koshino Y (1997) Effects of the 5-HT3 receptor agonist 1-(m-chlorophenyl)-biguanide in the rat kindling model of epilepsy. Brain Res 759(2):313–316. doi:10.1016/S0006-8993(97)00366-1
Yeetong P, Ausavarat S, Bhidayasiri R et al (2013) A newly identified locus for benign adult familial myoclonic epilepsy on chromosome 3q26.32-3q28. Eur J Hum Genet 21:225–228. doi:10.1038/ejhg.2012.133
Acknowledgments
We thank the families for their participation in this study. This work was supported by the National Health and Medical Research Council (NHMRC) Program Grant (628952) to J.G. M.B. was supported by an NHMRC Senior Research Fellowship (1002098) and NHMRC Program Grant (APP1054618). L.H. was supported by The John and Patricia Farrant Scholarship and the Australian Postgraduate Award Scholarship. This work was also supported by Victorian State Government Operational Infrastructure Support, Australian Government NHMRC IRIISS funding, Fondation Maladies rares, Assistance publique des hôpitaux de Paris (AP-HP) and Université Pierre et Marie-Curie (UPMC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Henden, L., Freytag, S., Afawi, Z. et al. Identity by descent fine mapping of familial adult myoclonus epilepsy (FAME) to 2p11.2–2q11.2. Hum Genet 135, 1117–1125 (2016). https://doi.org/10.1007/s00439-016-1700-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-016-1700-8