Abstract
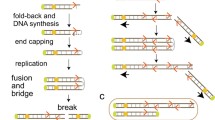
DNA amplification of the 11q13 region is observed frequently in many carcinomas. Within the amplified region several candidate oncogenes have been mapped, including cyclin D1, TAOS1 and cortactin. Yet, it is unknown which gene(s) is/are responsible for the selective pressure enabling amplicon formation. This is probably due to the use of low-resolution detection methods. Furthermore, the size and structure of the amplified 11q13 region is complex and consists of multiple amplicon cores that differ between different tumor types. We set out to test whether the borders of the 11q13 amplicon are restricted to regions that enable DNA breakage and subsequent amplification. A high-resolution array of the 11q13 region was generated to study the structure of the 11q13 amplicon and analyzed 29 laryngeal and pharyngeal carcinomas and nine cell lines with 11q13 amplification. We found that boundaries of the commonly amplified region were restricted to four segments. Three boundaries coincided with a syntenic breakpoint. Such regions have been suggested to be putatively fragile. Sequence comparisons revealed that the amplicon was flanked by two large low copy repeats known as segmental duplications. These segmental duplications might be responsible for the typical structure and size of the 11q13 amplicon. We hypothesize that the selection for genes through amplification of the 11q13.3 region is determined by the ability to form DNA breaks within specific regions and, consequently, results in large amplicons containing multiple genes.






Similar content being viewed by others
References
Albertson DG (2006) Gene amplification in cancer. Trends Genet 22(8):447–455
Armengol L, Pujana MA, Cheung J, Scherer SW, Estivill X (2003) Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum Mol Genet 12:2201–2208
Atayar C, Kok K, Kluiver J, Bosga A, van den Berg E, van d V, Blokzijl T, Harms G, Davelaar I, Sikkema-Raddatz B, Martin-Subero JI, Siebert R, Poppema S, van den Berg A (2006) BCL6 alternative breakpoint region break and homozygous deletion of 17q24 in the nodular lymphocyte predominance type of Hodgkin’s lymphoma-derived cell line DEV. Hum Pathol 37:675–683
Bailey JA, Eichler EE (2006) Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 7:552–564
Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE (2004) Hotspots of mammalian chromosomal evolution. Genome Biol 5:R23
Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Hoglund M, Johansson B, Hagemeijer A, Park SS, Mitelman F, Lupski JR, Fioretos T (2004) The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet 74:1–10
Barr FG, Nauta LE, Davis RJ, Schafer BW, Nycum LM, Biegel JA (1996) In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet 5:15–21
Ciullo M, Debily MA, Rozier L, Autiero M, Billault A, Mayau V, El Marhomy S, Guardiola J, Bernheim A, Coullin P, Piatier-Tonneau D, Debatisse M (2002) Initiation of the breakage-fusion-bridge mechanism through common fragile site activation in human breast cancer cells: the model of PIP gene duplication from a break at FRA7I. Hum Mol Genet 11:2887–2894
Coquelle A, Pipiras E, Toledo F, Buttin G, Debatisse M (1997) Expression of fragile sites triggers intrachromosomal mammalian gene amplification and sets boundaries to early amplicons. Cell 89:215–225
Coquelle A, Rozier L, Dutrillaux B, Debatisse M (2002) Induction of multiple double-strand breaks within an hsr by meganucleaseI-SceI expression or fragile site activation leads to formation of double minutes and other chromosomal rearrangements. Oncogene 21:7671–7679
Craig-Holmes AP, Strong LC, Goodacre A, Pathak S (1987) Variation in the expression of aphidicolin-induced fragile sites in human lymphocyte cultures. Hum Genet 76:134–137
De Gregori M, Pramparo T, Memo L, Gimelli G, Messa J, Rocchi M, Patricelli MG, Ciccone R, Giorda R, Zuffardi O (2005) Direct duplication 12p11.21–p13.31 mediated by segmental duplications: a new recurrent rearrangement? Hum Genet 118:207–213
Devriendt K, Matthijs G, Van Dael R, Gewillig M, Eyskens B, Hjalgrim H, Dolmer B, McGaughran J, Brondum-Nielsen K, Marynen P, Fryns JP, Vermeesch JR (1999) Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet 64:1119–1126
Difilippantonio MJ, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, Nussenzweig A (2002) Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med 196:469–480
Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP (2003) DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36:361–374
Freier K, Sticht C, Hofele C, Flechtenmacher C, Stange D, Puccio L, Toedt G, Radlwimmer B, Lichter P, Joos S (2006) Recurrent coamplification of cytoskeleton-associated genes EMS1 and SHANK2 with CCND1 in oral squamous cell carcinoma. Genes Chromosomes Cancer 45:118–125
Giglio S, Calvari V, Gregato G, Gimelli G, Camanini S, Giorda R, Ragusa A, Guerneri S, Selicorni A, Stumm M, Tonnies H, Ventura M, Zollino M, Neri G, Barber J, Wieczorek D, Rocchi M, Zuffardi O (2002) Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am J Hum Genet 71:276–285
Hagemeijer A, Lafage M, Mattei MG, Simonetti J, Smit E, de LO, Birnbaum D (1991) Localization of the HST/FGFK gene with regard to 11q13 chromosomal breakpoint and fragile site. Genes Chromosomes Cancer 3:210–214
Hermsen MA, Joenje H, Arwert F, Welters MJ, Braakhuis BJ, Bagnay M, Westerveld A, Slater R (1996) Centromeric breakage as a major cause of cytogenetic abnormalities in oral squamous cell carcinoma. Genes Chromosomes Cancer 15:1–9
Hermsen MA, Joenje H, Arwert F, Braakhuis BJ, Baak JP, Westerveld A, Slater R (1997) Assessment of chromosomal gains and losses in oral squamous cell carcinoma by comparative genomic hybridisation. Oral Oncol 33:414–418
Hermsen M, Guervos MA, Meijer G, Baak J, van Diest P, Marcos CA, Sampedro A (2001) New chromosomal regions with high-level amplifications in squamous cell carcinomas of the larynx and pharynx, identified by comparative genomic hybridization. J Pathol 194:177–182
Huang X, Gollin SM, Raja S, Godfrey TE (2002) High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci USA 99:11369–11374
Huang X, Godfrey TE, Gooding WE, McCarty KS Jr, Gollin SM (2006) Comprehensive genome and transcriptome analysis of the 11q13 amplicon in human oral cancer and synteny to the 7F5 amplicon in murine oral carcinoma. Genes Chromosomes Cancer 45:1058–1069
Hughes-Davies L, Huntsman D, Ruas M, Fuks F, Bye J, Chin SF, Milner J, Brown LA, Hsu F, Gilks B, Nielsen T, Schulzer M, Chia S, Ragaz J, Cahn A, Linger L, Ozdag H, Cattaneo E, Jordanova ES, Schuuring E, Yu DS, Venkitaraman A, Ponder B, Doherty A, Aparicio S, Bentley D, Theillet C, Ponting CP, Caldas C, Kouzarides T (2003) EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell 115:523–535
Hui AB, Or YY, Takano H, Tsang RK, To KF, Guan XY, Sham JS, Hung KW, Lam CN, van Hasselt CA, Kuo WL, Gray JW, Huang DP, Lo KW (2005) Array-based comparative genomic hybridization analysis identified cyclin D1 as a target oncogene at 11q13.3 in nasopharyngeal carcinoma. Cancer Res 65:8125–8133
Izzo JG, Papadimitrakopoulou VA, Li XQ, Ibarguen H, Lee JS, Ro JY, El Naggar A, Hong WK, Hittelman WN (1998) Dysregulated cyclin D1 expression early in head and neck tumorigenesis: in vivo evidence for an association with subsequent gene amplification. Oncogene 17:2313–2322
Janssen JW, Vaandrager JW, Heuser T, Jauch A, Kluin PM, Geelen E, Bergsagel PL, Kuehl WM, Drexler HG, Otsuki T, Bartram CR, Schuuring E (2000) Concurrent activation of a novel putative transforming gene, myeov, and cyclin D1 in a subset of multiple myeloma cell lines with t(11;14)(q13;q32). Blood 95:2691–2698
Jin Y, Hoglund M, Jin C, Martins C, Wennerberg J, Akervall J, Mandahl N, Mitelman F, Mertens F (1998) FISH characterization of head and neck carcinomas reveals that amplification of band 11q13 is associated with deletion of distal 11q. Genes Chromosomes Cancer 22:312–320
Jin Y, Jin C, Wennerberg J, Hoglund M, Mertens F (2002) Cyclin D1 amplification in chromosomal band 11q13 is associated with overrepresentation of 3q21–q29 in head and neck carcinomas. Int J Cancer 98:475–479
Jong K, Marchiori E, Meijer G, Van der Vaart A, Ylstra B (2004) Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics 20:3636–3637
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Kok K, Dijkhuizen T, Swart YE, Zorgdrager H, van d V, Fehrmann R, te Meerman GJ, Gerssen-Schoorl KB, van ET, Sikkema-Raddatz B, Buys CH (2005) Application of a comprehensive subtelomere array in clinical diagnosis of mental retardation. Eur J Med Genet 48:250–262
Kuo MT, Sen S, Hittelman WN, Hsu TC (1998) Chromosomal fragile sites and DNA amplification in drug-resistant cells. Biochem Pharmacol 56:7–13
Le Beau MM, Rassool FV, Neilly ME, Espinosa R, III, Glover TW, Smith DI, McKeithan TW (1998) Replication of a common fragile site, FRA3B, occurs late in S phase and is delayed further upon induction: implications for the mechanism of fragile site induction. Hum Mol Genet 7:755–761
Lin M, Smith LT, Smiraglia DJ, Kazhiyur-Mannar R, Lang JC, Schuller DE, Kornacker K, Wenger R, Plass C (2006) DNA copy number gains in head and neck squamous cell carcinoma. Oncogene 25:1424–1433
Lobachev KS, Gordenin DA, Resnick MA (2002) The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108:183–193
Locke DP, Segraves R, Nicholls RD, Schwartz S, Pinkel D, Albertson DG, Eichler EE (2004) BAC microarray analysis of 15q11–q13 rearrangements and the impact of segmental duplications. J Med Genet 41:175–182
Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422
Maser RS, DePinho RA (2002) Connecting chromosomes, crisis, and cancer. Science 297:565–569
Maser RS, DePinho RA (2004) Telomeres and the DNA damage response: why the fox is guarding the henhouse. DNA Repair (Amst) 3:979–988
Mills KD, Ferguson DO, Alt FW (2003) The role of DNA breaks in genomic instability and tumorigenesis. Immunol Rev 194:77–95
Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS (2006) The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125:1283–1296
Nelsen CJ, Kuriyama R, Hirsch B, Negron VC, Lingle WL, Goggin MM, Stanley MW, Albrecht JH (2005) Short term cyclin D1 overexpression induces centrosome amplification, mitotic spindle abnormalities, and aneuploidy. J Biol Chem 280:768–776
Newman T, Trask BJ (2003) Complex evolution of 7E olfactory receptor genes in segmental duplications. Genome Res 13:781–793
Nimeus E, Baldetorp B, Bendahl PO, Rennstam K, Wennerberg J, Akervall J, Ferno M (2004) Amplification of the cyclin D1 gene is associated with tumour subsite, DNA non-diploidy and high S-phase fraction in squamous cell carcinoma of the head and neck. Oral Oncol 40:624–629
Okuno Y, Hahn PJ, Gilbert DM (2004) Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res 32:749–756
Olender T, Feldmesser E, Atarot T, Eisenstein M, Lancet D (2004) The olfactory receptor universe–from whole genome analysis to structure and evolution. Genet Mol Res 3:545–553
Palakodeti A, Han Y, Jiang Y, Le Beau MM (2004) The role of late/slow replication of the FRA16D in common fragile site induction. Genes Chromosomes Cancer 39:71–76
Perucca-Lostanlen D, Hecht BK, Courseaux A, Grosgeorge J, Hecht F, Gaudray P (1997) Mapping FRA11A, a folate-sensitive fragile site in human chromosome band 11q13.3. Cytogenet Cell Genet 79:88–91
Pipiras E, Coquelle A, Bieth A, Debatisse M (1998) Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J 17:325–333
Reshmi SC, Gollin SM (2005) Chromosomal instability in oral cancer cells. J Dent Res 84:107–117
Richards RI (2001) Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet 17:339–345
Saunders WS, Shuster M, Huang X, Gharaibeh B, Enyenihi AH, Petersen I, Gollin SM (2000) Chromosomal instability and cytoskeletal defects in oral cancer cells. Proc Natl Acad Sci USA 97:303–308
Savelyeva L, Schwab M (2001) Amplification of oncogenes revisited: from expression profiling to clinical application. Cancer Lett 167:115–123
Schimke RT (1988) Gene amplification in cultured cells. J Biol Chem 263:5989–5992
Schuuring E (1995) The involvement of the chromosome 11q13 region in human malignancies: cyclin D1 and EMS1 are two new candidate oncogenes–a review. Gene 159:83–96
Schuuring E, van DH, Schuuring-Scholtes E, Verhoeven E, Michalides R, Geelen E, de BC, Brok H, van B, V, Kluin P (1998) Characterization of the EMS1 gene and its product, human Cortactin. Cell Adhes Commun 6:185–209
Schwab M (1999) Oncogene amplification in solid tumors. Semin Cancer Biol 9:319–325
Sen S, Hittelman WN, Teeter LD, Kuo MT (1989) Model for the formation of double minutes from prematurely condensed chromosomes of replicating micronuclei in drug-treated Chinese hamster ovary cells undergoing DNA amplification. Cancer Res 49:6731–6737
Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet 13 Spec No 1:R57–R64
Shuster MI, Han L, Le Beau MM, Davis E, Sawicki M, Lese CM, Park NH, Colicelli J, Gollin SM (2000) A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q13 amplification. Genes Chromosomes Cancer 28:153–163
Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG (2001) Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet 29:263–264
Stankiewicz P, Lupski JR (2002) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319
Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR (2004) Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res 14:2209–2220
Stark GR, Debatisse M, Giulotto E, Wahl GM (1989) Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57:901–908
Takes RP, Baatenburg de Jong RJ, Schuuring E, Hermans J, Vis AA, Litvinov SV, Van Krieken JH (1997) Markers for assessment of nodal metastasis in laryngeal carcinoma. Arch Otolaryngol Head Neck Surg 123:412–419
Tanaka H, Tapscott SJ, Trask BJ, Yao MC (2002) Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc Natl Acad Sci USA 99:8772–8777
Tchinda J, Dijkhuizen T, van der Vlies P, Kok K, Horst J (2004) Translocations involving 6p22 in acute myeloid leukaemia at relapse: breakpoint characterization using microarray-based comparative genomic hybridization. Br J Haematol 126:495–500
Toledo F, Le Roscouet D, Buttin G, Debatisse M (1992) Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO J 11:2665–2673
Toledo F, Coquelle A, Svetlova E, Debatisse M (2000) Enhanced flexibility and aphidicolin-induced DNA breaks near mammalian replication origins: implications for replicon mapping and chromosome fragility. Nucleic Acids Res 28:4805–4813
Trask BJ, Hamlin JL (1989) Early dihydrofolate reductase gene amplification events in CHO cells usually occur on the same chromosome arm as the original locus. Genes Dev 3:1913–1925
Vaandrager JW, Schuuring E, Zwikstra E, de Boer CJ, Kleiverda KK, Van Krieken JH, Kluin-Nelemans HC, van Ommen GJ, Raap AK, Kluin PM (1996) Direct visualization of dispersed 11q13 chromosomal translocations in mantle cell lymphoma by multicolor DNA fiber fluorescence in situ hybridization. Blood 88:1177–1182
van Dartel M., Hulsebos TJ (2004) Amplification and overexpression of genes in 17p11.2 ∼ p12 in osteosarcoma. Cancer Genet Cytogenet 153:77–80
Van Roy N., Vandesompele J, Menten B, Nilsson H, De SE, Rocchi M, De PA, Pahlman S, Speleman F (2006) Translocation-excision-deletion-amplification mechanism leading to nonsyntenic coamplification of MYC and ATBF1. Genes Chromosomes Cancer 45:107–117
Veltman JA, Schoenmakers EF, Eussen BH, Janssen I, Merkx G, van Cleef B, van Ravenswaaij CM, Brunner HG, Smeets D, van Kessel AG (2002) High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am J Hum Genet 70:1269–1276
Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B (2004) Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. Proc Natl Acad Sci USA 101:11368–11373
Watanabe Y, Fujiyama A, Ichiba Y, Hattori M, Yada T, Sakaki Y, Ikemura T (2002) Chromosome-wide assessment of replication timing for human chromosomes 11q and 21q: disease-related genes in timing-switch regions. Hum Mol Genet 11:13–21
Watanabe Y, Ikemura T, Sugimura H (2004) Amplicons on human chromosome 11q are located in the early/late-switch regions of replication timing. Genomics 84:796–805
Windle B, Draper BW, Yin YX, O’Gorman S, Wahl GM (1991) A central role for chromosome breakage in gene amplification, deletion formation, and amplicon integration. Genes Dev 5:160–174
Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP (2004) Replication timing of the human genome. Hum Mol Genet 13:575
Yuan B, Oechsli MN, Hendler FJ (1997) A region within murine chromosome 7F4, syntenic to the human 11q13 amplicon, is frequently amplified in 4NQO-induced oral cavity tumors. Oncogene 15:1161–1170
Yue Y, Haaf T (2006) 7E olfactory receptor gene clusters and evolutionary chromosome rearrangements. Cytogenet Genome Res 112:6–10
Acknowledgment
We would like to thank Trijnie Dijkhuizen, Pieter van der Vlies and Itty Oostendorp for technical assistance. We are grateful to Jane Briggs for reading the manuscript. This work is financially supported by the foundations of “Nijbakker-Morra”, “De Drie Lichten”, “Maurits en Anna de Kock” and the Groningen University Institute for Drug Exploration (GUIDE) in the Netherlands.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below are the links to the electronic supplementary material
Rights and permissions
About this article
Cite this article
Gibcus, J.H., Kok, K., Menkema, L. et al. High-resolution mapping identifies a commonly amplified 11q13.3 region containing multiple genes flanked by segmental duplications. Hum Genet 121, 187–201 (2007). https://doi.org/10.1007/s00439-006-0299-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00439-006-0299-6