Abstract.
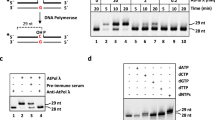
One of the major mutagenic base lesions in DNA caused by exposure to reactive oxygen species is 7,8-dihydro-8-oxoguanine (8-oxoG). Genes coding for DNA repair enzymes that recognise 8-oxoG have been reported in bacteria, yeast, mammals and plants. The prokaryotic and eukaryotic genes are functional homologues but differ in their primary sequence. We have cloned, sequenced, and expressed a new Arabidopsis thaliana cDNA that shows sequence homology to the eukaryotic genes coding for 8-oxoG DNA N-glycosylases (OGG1). The 40.3-kDa enzyme it encodes (AtOGG1) introduces a chain break in a double-stranded oligonucleotide specifically at an 8-oxoG residue. In addition, AtOGG1 can form a Schiff base with 8-oxoG in the presence of NaBH4, suggesting that it is a bifunctional DNA N-glycosylase. Furthermore, expression of AtOGG1 in an Escherichia coli strain that is deficient in the repair of 8-oxoG in DNA suppresses its spontaneous-mutator phenotype. Thus, we have demonstrated that AtOGG1 is not only a structural but also a functional eukaryotic OGG1 homologue.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Electronic Publication
Rights and permissions
About this article
Cite this article
Dany, A., Tissier, A. A functional OGG1 homologue from Arabidopsis thaliana . Mol Gen Genomics 265, 293–301 (2001). https://doi.org/10.1007/s004380000414
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s004380000414