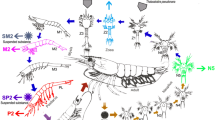
Abstract
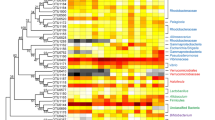
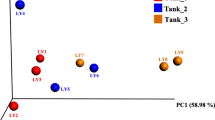
Gut microbiota is known to influence the physiology, health, nutrient absorption, reproduction, and other metabolic activities of aquatic organisms. Microbial composition can influence intestinal immunity and are considered as health indicators. Information on gut microbial composition provides potential application possibilities to improve shrimp health and production. In the absence of such information for Penaeus indicus, the present study reports the microbial community structure associated with its early developmental stages. Bacterial community associated with the early developmental stages (egg, nauplii, zoea, mysis, PL1, PL6 and PL12) from two hatchery cycles were analysed employing 16S rRNA high throughput sequencing. Proteobacteria and Bacteroidetes, were the two dominant phyla in P. indicus development stages. Sequential sampling revealed the constant change in the bacterial composition at genus level. Alteromonas was dominant in egg and nauplii stage, whilst Ascidiaceihabitans (formerly Roseobacter) was the dominant genera in both PL6 and PL12. The bacterial composition was highly dynamic in early stages and our study suggests that the mysis stage is the critical phase in transforming the microbial composition and it gets stabilised by early post larval stages. This is the first report on the composition of microbiota in early developmental stages of P. indicus. Based on these results the formation of microbial composition seems to be influenced by feeding at early stages. The study provides valuable information to device intervention strategies for healthy seed production.






Similar content being viewed by others
Availability of data and materials
The original contributions presented in the study are included in the article/Supplementary Information; further inquiries can be directed to the corresponding author.
Code availability
Not applicable.
References
Angthong P, Tanaporn U, Sopacha A, Panomkorn C, Nitsara K, Wanilada R (2020) Bacterial analysis in the early developmental stages of the black tiger shrimp (Penaeus monodon). Sci Rep 10:4896
Butt RL, Volkoff H (2019) Gut microbiota and energy homeostasis in fish. Front Endocrinol 10:9
Chen B, Du K, Sun C, Vimalanathan A, Liang X, Li Y, Wang B, Lu X, Li L, Shao Y (2018) Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J 12:2252–2262
Cheung MK, Yip HY, Nong W, Law PTW, Chu KH, Kwan HS, Hui JHL (2015) Rapid change of microbiota diversity in the gut but not the hepatopancreas during gonadal development of the new shrimp model Neocaridina denticulata. Mar Biotech 17:811–819
Crenn K, Duffieux D, Jeanthon C (2018) Bacterial epibiotic communities of ubiquitous and abundant marine diatoms are distinct in short- and long-term associations. Front Microbiol 9:2879–2879
Curiel FB, Ramirez-Pueblab ST, Ringøc E, Escobar-Zepedad A, Godoy-Lozanod E, Vazquez-Duhalte R, Sanchez-Floresd A, Vianaf MT (2018) Effects of extruded aquafeed on growth performance and gut microbiome of juvenile Totoaba macdonaldi. Anim Feed Sci Technol 245:91–103
Dhariwal A, Chong J, Habib S, King I, Agellon LB, Xia J (2017) MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45:180–188
FAO (2020) The state of world fisheries and aquaculture 2020. Sustainability in action. FAO, Rome. https://doi.org/10.4060/ca9229en
Hammer TJ, McMillan WO, Fierer N (2014) Metamorphosis of a butterfly-associated bacterial community. PLoS ONE 9:0086995
Hansen GH, Olafsen JA (1989) Bacterial colonization of Cod (Gadus morhua L.) and halibut (Hippoglossus hippoglossus) eggs in marine aquaculture. Appl Environ Microbiol 55:1435–1446
Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R (2015) InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinf 16:169
Holt CC, Bass D, Stentiford GD, Giezen MVD (2020) Understanding the role of the shrimp gut microbiome in health and disease. J Invert Pathol 21:107387
Johnson CN, Barnes S, Ogle J, Grimes DJ, Chang YJ, Peacock AD, Kline L (2008) Microbial community analysis of water, foregut, and hindgut during growth of Pacific white shrimp, Litopenaeus vannamei, in closed-system aquaculture. J World Aquac Soc 39:251–258
Kho ZY, Lal SK (2018) The human gut microbiome—a potential controller of wellness and diseases. Front Microbiol 9:1835
Klindworth A, Elmar P, Timmy S, Jorg P, Christian Q, Matthias H, Frank OG (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acid Res 4(1):11
Li E, Xu C, Wang X, Wang S, Zhao Q, Zhang Z, Qin GJ, Chen L (2018) Gut microbiota and its modulation for healthy farming of Pacific White Shrimp Litopenaeus vannamei. Rev Fish Sci Aquacult 26:381–399
Lim Y, Ilnam K, Cho JC (2020) Genome characteristics of Kordia antarctica IMCC3317T and comparative genome analysis of the genus Kordia. Sci Rep 10:14715
Liu H, Wang L, Liu M, Wang B, Jiang K, Ma S, Li Q (2011) The intestinal microbial diversity in Chinese shrimp (Fenneropenaeus chinensis) as determined by PCR–DGGE and clone library analyses. Aquaculture 317:32–36
Liu B, Bo L, Qunlan Z, Cunxin S, Changyou S, Huimin Z, Zhenfei Y, Shana F (2020) Patterns of bacterial community composition and diversity following the embryonic development stages of Macrobrachium rosenbergii. Aquacult Rep 17:100372
Luo H, Moran MA (2014) Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev 78(4):573–587
Meziti A, Mente E, Kormas KA (2012) Gut bacteria associated with different diets in reared Nephrops norvegicus. Syst Appl Microbiol 35:473–482
Naylor RL, Hardy RW, Buschmann AH, Bush SR, Cao L, Klinger DH, Little DC, Lubchenco J, Shumway SE, Max T (2021) A 20-year retrospective review of global aquaculture. Nature 591:551–563
Nelson TM, Rogers TL, Brown MV (2013) The gut bacterial community of mammals from marine and terrestrial habitats. PLoS ONE 8(12):e83655
Ooi MC, Goulden EF, Smith GG, Nowak BF, Bridle AR (2017) Developmental and gut-related changes to microbiomes of the cultured juvenile spiny lobster Panulirus ornatus. FEMS Microbiol Ecol 93:fix159
Patil PK, Vinay TN, Ghate SD, Baskaran V, Avunje S (2021) 16S rRNA gene diversity and gut microbial composition of the Indian white shrimp (Penaeus indicus). Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-021-01658-9
Rajeev R, Adithya KK, Kiran GS, Selvin J (2021) Healthy microbiome: a key to successful and sustainable shrimp aquaculture. Rev Aquacult 13:238–258
Rungrassamee W, Klanchui A, Chaiyapechara S, Maibunkaew S, Tangphatsornruang S (2013) Bacterial population in intestines of the Black Tiger Shrimp (Penaeus monodon) under different growth stages. PLoS ONE 8(4):e60802
Rungrassamee W, Klanchui A, Maibunkaew S, Chaiyapechara S, Jiravanichpaisal P, Karoonuthaisiri N (2014) Characterization of intestinal bacteria in wild and domesticated adult black tiger shrimp (Penaeus monodon). PLoS ONE 9(3):e91853
Rungrassamee W, Klanchui A, Maibunkaew S, Karoonuthaisiri N (2016) Bacterial dynamics in intestines of the black tiger shrimp and the Pacific white shrimp during Vibrio harveyi exposure. J Invert Pathol 133:12–19
Sajeela KA, Gopalakrishnan A, Basheer VS, Mandal A, Bineesh KK, Grinson G, Gopakumar SD (2019) New insights from nuclear and mitochondrial markers on the genetic diversity and structure of the Indian white shrimp Fenneropenaeus indicus among the marginal seas in the Indian Ocean. Mol Phylogenet Evol 136:53–64
Schloss PD, Westcott SL, Raybin T, Hall JR, Hartmann M (2009) Introducing Mothur: open-source. platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schmidt VT, Smith KF, Melvin DW, Amaral-Zettler LA (2015) Community assembly of a euryhaline fish microbiome during salinity acclimation. Mol Ecol 24:2537–2550
Stephens WZ, Burns AR, Keaton S, Sandi W, John FR, Karen G, Brendan JMB (2016) The composition of the zebrafish intestinal microbial community varies across development. ISME J 10:644–654
Turnes MSG, Hay ME, Fenical W (1989) Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246(4926):116–118
Vijayan KK (2019) Domestication and genetic improvement of Indian white shrimp, Penaeus indicus: a complimentary native option to exotic Penaeus vannamei. J Coast Res 86:270–276
Wang H, Huang J, Wang P, Li T (2020) Insights into the microbiota of larval and postlarval Pacific white shrimp (Penaeus vannamei) along early developmental stages: a case in pond level. Mol Genet Genom 295:1517–1528
Yao Z, Yang K, Huang L, Huang X, Qiuqian L, Wang L, Zhang D (2018) Disease outbreak accompanies the dispersive structure of shrimp gut bacterial community with a simple core microbiota. AMB Expr 8:120
Zhang Z, Liu J, Jin X, Liu C, Fan C, Guo L, Liang Y, Zheng J, Peng N (2020) Developmental, dietary, and geographical impacts on gut microbiota of red swamp crayfish (Procambarus clarkii). Microorganisms 8:1376
Zheng Y, Min Y, Liu J, Qiao Y, Wang L, Li Z, Zhang XH, Yu M (2017) Bacterial community associated with healthy and diseased Pacific white shrimp (Litopenaeus vannamei) larvae and rearing water across different growth stages. Front Microbiol 8:1362
Zheng D, Liwinski T, Elinav E (2020) Interaction between microbiota and immunity in health and disease. Cell Res 30:492–506
Acknowledgements
Authors acknowledge the facilities and support provided by the Director, ICAR-CIBA to carry out this work through the institute funded project FISHCIBASIL201800200130 & Consortia Research Platform (CRP) on Vaccine and Diagnostics (CRP-V&D) 1007611.
Author information
Authors and Affiliations
Contributions
PKP and VTN: planning of the study, PKP: critical evaluation and fund mobility. VTN, AR, and SAPS: shrimp seed production and larval rearing, VTN and VB: sampling and Sequencing. VTN: Metagenomic analysis and data interpretation. PKP and VTN wrote the manuscript, CPB: reviewed and provided valuable inputs to the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Martine Collart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Figure S1
Linear discriminative analysis (LDA) effect size (LEfSe) of potential taxonomic biomarkers among different developmental stages of P. indicus. 34 genera, varying significantly were identified with LDA scores >4.. Supplementary file1 (PDF 17 KB)
Rights and permissions
About this article
Cite this article
Vinay, T.N., Patil, P.K., Aravind, R. et al. Microbial community composition associated with early developmental stages of the Indian white shrimp, Penaeus indicus. Mol Genet Genomics 297, 495–505 (2022). https://doi.org/10.1007/s00438-022-01865-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-022-01865-7