Abstract
Interspecific introgression can occur between species that evolve rapidly within an adaptive radiation. Pachyptila petrels differ in bill size and are characterised by incomplete reproductive isolation, leading to interspecific gene flow. Salvin’s prion (Pachyptila salvini), whose bill width is intermediate between broad-billed (P. vittata) and Antarctic (P. desolata) prions, evolved through homoploid hybrid speciation. MacGillivray’s prion (P. macgillivrayi), known from a single population on St Paul (Indian Ocean), has a bill width intermediate between salvini and vittata and could also be the product of interspecies introgression or hybrid speciation. Recently, another prion population phenotypically similar to macgillivrayi was discovered on Gough (Atlantic Ocean), where it breeds 3 months later than vittata. The similarity in bill width between the medium-billed birds on Gough and macgillivrayi suggest that they could be closely related. In this study, we used genetic and morphological data to infer the phylogenetic position and evolutionary history of P. macgillivrayi and the Gough medium-billed prion relative other Pachyptila taxa, to determine whether species with medium bill widths evolved through common ancestry or convergence. We found that Gough medium-billed prions belong to the same evolutionary lineage as macgillivrayi, representing a new population of MacGillivray’s prion that originated through a colonisation event from St Paul. We show that macgillivrayi’s medium bill width evolved through divergence (genetic drift) and independently from that of salvini, which evolved through hybridisation (gene flow). This represents the independent convergence towards a similarly medium-billed phenotype. The newly discovered MacGillivray’s prion population on Gough is of utmost conservation relevance, as the relict macgillivrayi population in the Indian Ocean is very small.
Similar content being viewed by others
Introduction
Mayr (1963) affirmed that ‘‘Without speciation there would be no diversification of the organic world, no adaptive radiation, and very little evolutionary progress’’. Thus, investigating how species evolve is crucial to understand the evolutionary processes on Earth and the influence of speciation on both species’ persistence and patterns of species diversity (Seehausen et al. 2014). Given current Anthropocene biodiversity crisis, this could help develop effective ways to protect existing diversity and the diversification process itself (Rosenzweig 2001). A defining feature of species is that their populations are connected and integrity is maintained by gene flow (Rieseberg et al. 2004; Petit and Excoffier 2009), whereas reproductive barriers between species are important as they preserve adaptations (Mayr 1942). However, reproductive barriers are likely to be semipermeable to gene flow in recently diverged species or taxa in which hybrid incompatibilities evolve slowly, and thus speciation can occur in the presence interspecific gene flow (introgression) (Rieseberg et al. 2004; Seehausen et al. 2014). In line with this, Mayr’s (1963) notion that gene flow counteracts the process of speciation in animals is being eroded by examples of interspecific introgression, which can sometimes be adaptive (e.g. Heliconius butterflies, Nadeau et al. 2012), or occasional cases of homoploid hybrid speciation, where a fully reproductively isolated species can evolve if hybrid fitness is high (Nolte et al. 2006; Kunte et al. 2011; Lamichhaney et al. 2018). In most instances, interspecific introgression occurs between species that have diverged recently within the same adaptive radiation, because although such species have evolved distinct phenotypic traits, they are still sufficiently closely related to produce viable offspring (Seehausen et al. 2014; Masello et al. 2019).
One such adaptive radiation is that of the prions, Pachyptila, a genus of small petrels (Procellariidae) that differ primarily in bill size and structure (Warham 1990). Despite large oceanic distances separating breeding colonies, the prions are characterised by high levels of gene flow and incomplete reproductive isolation (Masello et al. 2019), which has blurred species limits resulting in the recognition of anything from three to nine Pachyptila species (Mathews 1934; Murphy 1936; Falla 1940; Fleming 1941; Fullagar 1972; Cox 1980; Harper 1980; Bretagnolle et al. 1990; Penhallurick and Wink 2004; Rheindt and Austin 2005; Howell and Zufelt 2019). Traditionally, most confusion has surrounded the larger-billed taxa (broad-billed prion P. vittata, average bill width, 21.4 mm, Salvin’s prion P. salvini, 17.1 mm, and Antarctic prion P. desolata, 14.3 mm), which have palatal lamellae on their upper mandibles adapted to filter zooplankton (Masello et al. 2019). The number of palatal lamellae is correlated with bill width, and these two variables determine the range of species that can be preyed upon (Masello et al. 2019). Palatal lamellae are best developed in P. vittata, allowing it to feed almost exclusively on copepods, whereas P. desolata, which feeds primarily on hyperiid amphipods, has less well-developed lamellae (Imber 1981; Bretagnolle et al. 1990; Klages and Cooper 1992; Cherel et al. 2002). The narrowest billed species (thin-billed prion P. belcheri, average bill width, 11 mm, fairy prion P. turtur, 11 mm; Masello et al. 2019) have only vestigial palatal lamellae and do not filter feed (Murphy 1936; Morgan and Ritz 1982; Bretagnolle et al. 1990; Klages and Cooper 1992). Since the larger-billed prions do not form a monophyletic clade, and all species except P. belcheri and P. turtur possess functional palatal lamellae, the most parsimonious explanation would be that both broad bills and lamellae represent the ancestral state in prions, with lamellae becoming vestigial once bill width reduced to below 12 mm (Masello et al. 2019). Murphy (1936) suggested that the thin bill of P. belcheri was structurally ancestral to that of P. desolata; however, the ancestral state of bill width in prions remains to be investigated. Genetically, Pachyptila comprises two distinct evolutionary clades, the first comprising the widest-billed species, P. vittata, and the other containing all other species (P. turtur, P. belcheri, P. desolata, and P. salvini) (Masello et al. 2019).
Pachyptila salvini, whose bill width is on average intermediate (17.1 mm) between P. vittata (21.4 mm) and P. desolata (14.3 mm), has the ability to feed on both copepods and hyperiid amphipods (Gartshore and Steele 1988; Ridoux 1994; Masello et al. 2019). Furthermore, although mitochondrial DNA (mtDNA) places P. salvini within the narrower-billed clade, coalescent simulations of microsatellite DNA showed that P. salvini evolved in a rare case of homoploid hybrid speciation between P. vittata and P. desolata (Masello et al. 2019). Pachyptila salvini’s intermediate bill width, with respect to P. vittata and P. desolata, allows it to feed on more prey species, giving it a potential feeding advantage over either of its parental species (Bretagnolle et al. 1990; Masello et al. 2019). Remarkably, P. salvini’s mid-summer breeding time is also intermediate between that of P. vittata (early summer) and P. desolata (late summer), thus isolating it reproductively, which might otherwise have led to the disappearance of its hybrid phenotype.
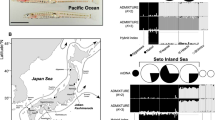
MacGillivray’s prion (P. macgillivrayi) is another taxon with an average bill width (17.3 mm) intermediate between P. salvini and P. vittata (Roux et al. 1986). Although several authors recognise P. macgillivrayi as a species in its own right (Bretagnolle et al. 1990; Worthy and Jouventin 1999; Shirihai 2007), others have regarded it as a subspecies of either P. salvini (Roux et al. 1986) or P. vittata (Harper 1980). This little-known taxon comprises a single relict population confined to St Paul Island (38.7° S) in the southern Indian Ocean (Tollu 1984; Micol and Jouventin 2002; Jiguet et al. 2007), but it was formerly abundant on nearby Amsterdam Island (37.8° S, Worthy and Jouventin 1999; Fig. 1). Pachyptila macgillivrayi samples have been unavailable until recently, so the species was not included in the evolutionary genetic analysis of Masello et al. (2019). Its phylogenetic position with respect to narrower- and broader-billed clades (sensu Masello et al. 2019) and its microsatellite DNA allelic distribution are therefore unknown. Could the intermediate bill width of P. macgillivrayi also be a product of interspecies introgression, or even hybrid speciation?
Intriguingly, another prion population which is phenotypically similar to P. macgillivrayi was recently discovered on Gough Island in the South Atlantic Ocean, at similar latitude (40.3° S) to St Paul and Amsterdam islands in the Indian Ocean (Ryan et al. 2014). Gough Island is also home to the largest colony of P. vittata, which has disjunct South Atlantic (Tristan da Cunha and Gough) and southwest Pacific Ocean (New Zealand) populations (Fig. 1). On Gough Island, the recently discovered birds (hereafter Gough medium-billed) partially overlap in breeding distribution with P. vittata, but show a marked difference in bill width, with an average (18.5 mm; Ryan et al. 2014) that is also intermediate between P. desolata (14.3 mm) and P. vittata (21.4 mm; Masello et al. 2019). Given its similarity in bill width with P. salvini (17.1 mm) and P. macgillivrayi (17.3 mm; Roux et al. 1986; Masello et al. 2019), the Gough medium-billed population could be related to either species or represent a novel lineage. The Gough medium-billed prions breed roughly 3 months later than P. vittata, suggesting that they are a separate species (Ryan et al. 2014). Jones et al. (2020) showed that tracked P. vittata and Gough medium-billed individuals foraged and moulted in different areas of the South Atlantic Ocean, with the latter generally occurring farther south than P. vittata. Both the temporal and the latitudinal segregation have also been found in other prion species breeding in sympatry (Bretagnolle et al. 1990; Quillfeldt et al. 2015, 2020).
In this study, we report a morpho-genetic survey of P. macgillivrayi and both Pachyptila populations on Gough Island, and combine this with existing comparative data for other prion species to infer the phylogenetic position and evolutionary history of the Gough medium-billed prion, P. macgillivrayi and other Pachyptila taxa, given the poorly developed reproductive isolation in this genus, and to determine the ancestral state of bill width in prions.
Materials and methods
Samples and morphometric data
We first performed an analysis of average bill widths across populations of the larger-billed Pachyptila species, including Gough medium-billed (Ryan et al. 2014). We measured maximum bill widths of live or freshly dead adult prions to the nearest 0.1 mm on islands in the Tristan da Cunha archipelago (P. vittata), Gough Island (P. vittata and Gough medium-billed), St Paul Island (P. macgillivrayi), Marion Island (P. salvini), and beached specimens from South Africa (P. desolata; Fig. 1, for sample sizes see Fig. 2). We investigated differences in bill width using the Kruskal–Wallis test and Dunn's homogenous subgroups implemented in R (R Development Core Team 2019), as normality and equality of variance were not satisfied (p < 0.05).
Differences in bill widths of adult larger-billed prions (Pachyptila). Box plot, Dunn’s homogenous subgroups (a–e), and sample sizes of the populations measured in this study. Box plots include medians, whiskers indicate variability outside the upper and lower quartiles (grey shaded block), and circles are outliers. Data from Masello et al. (2019) and references therein are shown here in green for comparison purposes; green triangles represent means for different colonies (turtur: 7; belcheri: 3; desolata: 7; salvini: 2; vittata: 1)
For DNA analysis we collected muscle samples of P. macgillivrayi on St Paul Island (Fig. 1, Table 1). In addition, we sampled subfossil bones of P. macgillivrayi from Amsterdam Island (Table 1; Museum of New Zealand Te Papa Tongarewa registration numbers and GenBank accession numbers in Supplemental Material, Table S1). We also sampled both Gough medium-billed as well as P. vittata on Gough Island (Fig. 1, Table 1). To ensure we sequenced individuals from the correct populations, samples from Gough were taken from birds found outside the range of morphological overlap, which is roughly a bill width between 19.5 and 20.5 mm (Ryan et al. 2014). Therefore, all P. vittata sampled had bills > 21 mm wide and were collected from the area around the meteorological station where P. vittata predominates, and all Gough medium-billed birds had bills < 19 mm and were collected from Gonydale, a highland valley where medium-billed birds occur. To place our newly sampled populations in broader context, we compared their DNA data to previously sequenced or genotyped samples (Masello et al. 2019), and to new samples from other taxa. These additional samples were collected from birds at different breeding localities (Fig. 1) between 1999 and 2014, including P. vittata from Tristan da Cunha (Nightingale Island), islets off Stewart Island, Snares Island and the Chatham Islands (Rangatira Island) off New Zealand; P. salvini from the Prince Edward Islands (Marion Island) and the Crozet Islands; P. desolata from Kerguelen, South Georgia and Macquarie Island; P. turtur from Mana Island (New Zealand), and the closely related blue petrel (bird wrecked at New Zealand) to be used as outgroup (Table 1). Most DNA samples were extracted from fragments of muscle or skin tissue collected from fresh prion corpses, but some feathers and blood samples were taken from live birds (Table 1). Tissue samples were stored in ethanol, dry as feather quills, blood in Queens’s lysis buffer (Kerguelen) and blood on FTA classic cards (Whatman International Ltd, Maidstone, UK; Chathams, South Georgia, Macquarie). Some carcasses were stored frozen prior to sampling. Samples sizes are provided in Table 1.
Molecular methods
For the subfossil bones a ~ 5 mm fragment of bone was removed from the broken ends of humeri using a Dremel grinder with a new Dremel wheel used for each bone. Bone samples were powdered by grinding in sterilised mortars and pestles. Bone powder was then decalcified and a phenol–chloroform extraction performed (Shepherd and Lambert 2008) to isolate the DNA.
DNA was extracted from modern samples using a Qiagen DNeasy® Blood and Tissue kit (Qiagen, Germany), following the manufacturer’s instructions. The final elution volume was 100 µl for blood and modern tissue extractions and 45 µl for feather samples. Extractions and PCR setups from subfossil bones were performed in a dedicated ancient DNA (aDNA) laboratory located in a different building from where modern DNA and PCR products were handled. Potential contamination was monitored by the use of negative extraction and PCR controls.
The mitochondrial cytochrome b (cyt b) gene has previously provided a reasonable estimate of maternal evolutionary relationships among a range of Pachyptila species, which were largely congruent with data from nuclear microsatellites (Moodley et al. 2015) and nuclear introns (Masello et al. 2019). We therefore used this marker, as well as sequence data from the cytochrome c oxidase subunit 1 (COI), to reconstruct maternal relationships among these taxa using a subset of 44 samples (Table 1). An 811-base pair (bp) fragment of cyt b was PCR amplified using specific primers (CytB_Pri_F: 5′-CTAGCTATACACTACACCGC-3′ and CytB_Pri_R: 5′-CTAGTTGGCCGATGATGATG-3′) (Moodley et al. 2015). Primer3 (Untergasser et al. 2012) was used to design novel internal primers for amplifying cyt b in ancient DNA samples. These primers amplified five overlapping fragments which ranged in size from 209 to 269 bp (CytB_Pri_F with Pricytbint1R: 5′-AGGATGACTCCTGTGTTTCATGT-3′; Pricytbint2F: 5′-CCACATTGGACGAGGACTTT-3′ with Pricytbint2R: 5′-GGCAAAGAATCGGGTTAGTG-3′; Pricytbint3F: 5′-CCCTCGTAGAATGAGCCTGA-3′ with Pricytbint3R: 5′- GGGGGAGAATAGGGCTAAAG-3′; Pricytbint4F: 5′- CGGCATCGTATCAAACTGTG-3′ with Pricytbint4R: 5′- TTGAGCGTAGGATGGCATAA-3′ and Pricytbint5F: 5′-YCCTCCCCATATTAAACCAGA-3′ with CytB_Pri_R). A 648 bp fragment of cytochrome c oxidase subunit I (COI) was amplified from the modern samples using the AWCF1 and AWCR6 primers of Patel et al. (2010). Ancient DNA samples were amplified and sequenced from short overlapping fragments of COI using internal primers from Patel et al. (2010).
PCR amplifications of cyt b were performed in 20 μl reaction volumes containing 100 ng DNA template, 10 mM of each primer, 10 mM dNTPs (Roth, Karlsruhe), 2 mM MgCl and 5 U Taq DNA polymerase (BioLabs Taq DNA polymerase) in a 1 × PCR reaction buffer. Thermocycling conditions involved an initial denaturation at 94 °C for 2 min, 30 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 45 s and extension at 72 °C for 1 min, followed by a final extension step of 5 min at 72 °C. PCRs for COI were conducted in 10 μl volumes containing 1 × PCR buffer, 200 μM of dNTP, 0.5 U of Taq DNA polymerase (Roche), 0.3 M of BSA and 0.5 µM of primer. Thermocycling involved initial denaturation of 2 min at 94 °C, followed by 35 cycles of 94 °C for 30 s, 50 °C for 40 s and 72 °C for 1 min, followed by a final extension of 10 min at 72 °C.
PCR products were purified by digestion with exonuclease-shrimp alkaline phosphatase (from USB Corp, Cleveland for COI and Fermentas Life Sciences for cyt b), following the manufacturer’s specifications. PCR products were then sequenced in both directions using Big Dye chemistry (Applied Biosystems) and run on an AB 3130xl genetic analyser (for cyt b) or ABI3730 (for COI). Cyt b sequences were assembled and aligned in CLC Main Workbench 6.9.2 (CLC bio, Aarhus, Denmark). COI sequences were edited in Sequencer 5.2.3 (Gene Codes Corporation), and were aligned manually as they contained no indels.
Twenty-five previously isolated prion microsatellite loci (Moodley et al. 2015) were also amplified from genomic DNA of the St Paul P. macgillivrayi and Gough medium-billed samples. These were run together with samples of known genotype, so that these data may be calibrated against those of other prion microsatellite studies. Microsatellite profiles were checked for null alleles using MICROCHECKER (van Oosterhout et al. 2004) and for deviation from genotypic equilibrium (Hardy Weinberg equilibrium, HWE) using FSTAT (Goudet 1995). Multiple tests were corrected for using a Bonferroni correction. For context, these newly generated microsatellite data were analysed together with 216 samples of P. vittata from Gough, Tristan and Chathams, P. salvini from Marion, and P. desolata from Kerguelen, South Georgia and Macquarie (Table 1), published in previous studies (Moodley et al. 2015; Quillfeldt et al. 2017; Masello et al. 2019).
Genetic diversity and structure
Genetic diversity parameters were estimated for cyt b and COI for each population with at least five samples using DnaSP v5 (Librado and Rozas 2009) (Table 2). We estimated also two demographic indicators, Tajima’s D (1989) and Fu’s Fs (1997) (Table 2), for each gene and each species/population using DnaSP v5. For microsatellites, the allelic richness (AR, mean number of alleles per locus) was estimated using GENETIX 4.05 (Belkhir et al. 2004) and rarefied for differences in sample size using ADZE 1.0 (Szpiech et al. 2008). Unbiased expected heterozygosity (HE) and observed heterozygosity (HO) were also estimated in GENETIX.
Maternal genetic structure among prion species was determined using two methods. First, we concatenated both mtDNA genes and generated a median-joining network (Bandelt et al. 1999) of the composite haplotypes in POPART (Leigh and Bryant 2015). Then, we used the same DNA sequence data to reconstruct phylogenetic relationships using a Bayesian multispecies coalescent framework in BEAST 2 (Bouckaert et al. 2014). The best nucleotide substitution model was determined for each of the two alignments using JMODELTEST (Posada 2008). Site models were thus unlinked for each partition. Each gene tree was unlinked, and a species tree using a birth–death model prior was used to account for gene tree incongruence. The population prior allowed for changes in effective population size, but assumed a constant ancestral population size. All clock models were unlinked, but to determine the correct clock prior, alternative models assuming lognormal and exponential priors were tested against a strict molecular clock for each partition. A soft-bounded lognormal prior (α = 1.2, β = 1.0) of no later than 5 million years for the emergence of the genus was based on the earliest fossil evidence (Olson 1983, 1985a, b). After a single run of 100,000,000 MCMC iterations, sampling every 100,000 steps and discarding 20% as burn-in, the standard deviations of the posterior marginal distributions of both these parameters included zero in all cases, making them no more likely than a null strict clock for all gene partitions. All subsequent analyses were therefore carried out assuming a strict clock. Since we were unsure of how P. macgillivrayi and Gough medium-billed population were related to other prions, we also included two samples of taxa with vestigial lamellae (P. belcheri and P. turtur), as well as H. caerulea as outgroup (Table 1), to help identify the narrower-billed clade in both network and phylogenetic analyses.
Nuclear genetic structure was assessed using the Bayesian clustering algorithm implemented in STRUCTURE 2.3.4 (Pritchard et al. 2000). We assumed an admixture model because our previous molecular analysis of prions (Masello et al. 2019) showed consistent evidence of incomplete reproductive isolation and secondary contact. Moreover, inter-island movements have been recorded among well-studied procellariiforms (e.g. wandering albatross Diomedea exulans; Inchausti and Weimerskirch 2002), as well as a lack of genetic structure in many oceanic pelagic species in the Southern Ocean (e.g. Burg and Croxall 2001), including some Atlantic and Indian Ocean Pachyptila populations (Quillfeldt et al. 2017). The analysis was run ten times for K1–10 with each run randomly started, consisting of 500,000 Markov Chain Monte Carlo (MCMC) iterations, assuming correlated allele frequencies and removing the first 100,000 runs as burn-in. The mean likelihood values across multiple values of K were determined using STRUCTURE HARVESTER (Earl and vonHoldt 2012) in accordance with Evanno’s method (ΔK, i.e. the rate of change in the log probability of data between successive K values) (Evanno et al. 2005). Assignment plots were constructed for all values of K that were biologically interpretable. We used CLUMPAK (Kopelman et al. 2015) for the creation of genetic ancestry figures. We also used ARLEQUIN 3.5 (Excoffier and Lischer 2010) to implement analyses of molecular variance (AMOVAs) on the microsatellite data. We tested several a priori groupings of populations, with the expectation that the variance component distributed between populations (FST) and among defined groups (Fct) would be highest for the evolutionarily correct grouping. The significance of F statistics and variance components were tested with 99,999 permutations. We also calculated pairwise FST (after Weir and Cockerham 1984) between all populations in ARLEQUIN 3.5 (Excoffier and Lischer 2010), with significance again determined with 99,999 permutations.
Given previously observed levels of introgression in prion species (Masello et al. 2019), we determined the proportion of genetic variation within P. macgillivrayi and Gough medium-billed prions that was derived through introgression using Bayesian inference. Since we were only interested in gene flow into and out of the two newly sampled medium-billed populations, we tested for the presence of bi-directional migration between P. macgillivrayi and Gough medium-bills, as well as between both these populations and Gough’s sympatric P. vittata population. We inferred gene flow using BAYESASS 3.0 (Wilson and Rannala 2003), which estimates the posterior probability of an individual’s history and allows an estimation of the rate and direction of recent dispersal (Genovart et al. 2013). The acceptance rates for the main parameters (i.e. ‘migration’ rate, inbreeding coefficient and allele frequencies) were adjusted during preliminary runs. Convergence was evaluated by inspection of the trace files in TRACER 1.5 (Rambaut et al. 2018). Final parameter estimates were attained after performing three independent runs by means of different starting random seed numbers. The MCMC was run for 50,000,000 iterations with a burn-in period of 10,000,000 and a sampling frequency of 5000 iterations.
Reconstruction of ancestral bill state
To understand the evolution of bill width among Pachyptila species, we reconstructed the ancestral state of bill width using BAYESTRAITS v3.0.2 (Pagel et al. 2004). We used bill width data collected in the present study for larger-billed species, together with mean bill width of P. turtur and P. belcheri from Masello et al. (2019), and the phylogenetic tree (topology and branch lengths) reconstructed above. We also measured bill width of blue petrels Halobaena caerulea from South Georgia and Kerguelen (Supplemental Material, Table S2). Since this trait varied continuously among taxa, we selected the “Continuous: Random Walk” option (Pagel 1999, Model A), which uses a generalised least squares model to reconstruct the posterior distribution of bill width at the ancestral node of the Pachyptila tree. We accounted for phylogenetic uncertainty by using the entire post-burn-in posterior sample of trees generated by BEAST 2. We then inferred posterior distribution of ancestral bill widths using MCMC iterations, with a chain length of 1,001,000 iterations and a burn-in of 200,200 iterations. We performed this analysis with and without the outgroup H. caerulea to examine whether the ancestral state of bill width changed since the genera Pachyptila and Halobaena shared a common ancestor.
Additional comparisons
We also report variation in bill colour among the prions at Tristan and Gough, to investigate its usefulness for distinguishing among Pachyptila taxa, given the assertion that P. vittata differs from all other prions in having a blackish bill (e.g. Marchant and Higgins 1990; Shirihai 2007).
Results
Bill width varied significantly among the larger-billed prion taxa measured in this study (Kruskal–Wallis, χ2 = 679.5, df = 5, P ≤ 0.001). An average bill width of 14.2 mm was found in P. desolata, 16.7 mm in P. salvini, 18.0 mm for both Gough medium-billed and P. macgillivrayi, and 21.8 and 21.4 mm for P. vittata from Gough and Tristan, respectively (Fig. 2, and Supplemental Material, Table S2). Dunn’s homogenous subgroups revealed that the Gough medium-billed prions were similar to P. macgillivrayi (homogeneous subgroup ‘c’), and that both taxa were significantly different from all other prion taxa (Fig. 2, and Supplemental Material, Table S3).
The final mtDNA alignment, including outgroup sequences, comprised 811 bp of cyt b sequence and 648 bp of COI sequence from 47 individuals. This alignment also included a sequence from a subfossil P. macgillivrayi bone from the extinct Amsterdam Island population (S.34710.1) that produced full-length COI and cyt b sequences (Table 1, and Supplemental Material, Table S1). The COI sequence obtained for this sample was identical to the most common haplotype detected from modern P. macgillivrayi sampled on St Paul Island and the cyt b sequence differed from the most-closely related sequence by two substitutions. Only a 231 bp COI sequence could be obtained from the Amsterdam Island P. macgillivrayi bone specimen S.35077.1 and this was identical to the sequence from S.34710.1 and was not used in further analyses (Supplemental Material, Table S1).
Genetic diversity was higher for COI than for cyt b (Table 2), with P. macgillivrayi also featuring a higher relative diversity at the former locus and a lower diversity at the latter. Both loci showed negative values for Tajima’s D and Fu’s Fs which, assuming mitochondrial neutrality, may indicate signatures of recent population expansions, although only the P. macgillivrayi population from St Paul Island showed significantly negative Tajima’s D for COI (Table 2). Average microsatellite diversity indices showed equally high values for the investigated populations, with relatively higher values for P. vittata from Gough Island (Table 2). Private allelic richness was highest among the Gough medium-billed prions but, in general, was relatively low (Table 2).
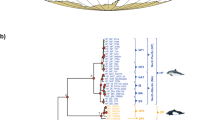
The maternal phylogenetic relationships among prion taxa inferred from the median-joining haplotype networks and the Bayesian species tree (Fig. 3) identified the two previously detected broader- and narrower-billed clades (Masello et al. 2019), with the narrow-billed P. turtur diverging basally some 5–7.1 Mya (95% highest posterior density, HPD). Pachyptila macgillivrayi samples from St Paul and Amsterdam Island formed a monophyletic clade. This clade was sister to another monophyletic clade made up exclusively of Gough medium-billed prions, which appears to have diverged from other P. macgillivrayi about 0.8–3.3 Mya (95% HPD, Fig. 3B). Interestingly, this monophyletic P. macgillivrayi-Gough medium-billed clade was not closely related to P. salvini, as was expected from their similar intermediate bill widths (Fig. 2), instead clustering within the broad-billed evolutionary group, sister to P. vittata (Fig. 3B).
Medium-joining haplotype network (A) and inferred Bayesian phylogeny (B) based on the combined sequence data for cyt b and COI for the prion taxa (Pachyptila) in this study. In the network, the circle size is proportional to haplotype frequency, hash marks crossing line connections represent mutational steps, and nodes without circles correspond to hypothetical haplotypes not sampled. In the inferred phylogeny, for clarity reasons, 95% HPD bars and probabilities are only shown for nodes with posterior probabilities > 0.5
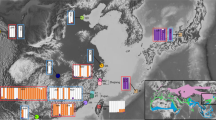
This mtDNA structure was broadly reflected in the microsatellite data (Fig. 4 and Supplemental Material, Fig. S1 for K1–10). After STRUCTURE analysis, the Evanno method (Evanno et al. 2005) estimated the highest ΔK (101.5) for K = 2, in which narrower- and broader-billed groups were separated. In contrast to mtDNA, P. macgillivrayi and Gough medium-billed prions partitioned with narrower-billed species at K = 2, however, at K = 3, both formed their own cluster, distinct from both narrower- and broader-billed groups. Structure analyses also revealed that the multilocus allele profiles of P. vittata from Gough appear admixed with P. macgillivrayi alleles (Fig. 4).
Genetic ancestry based on an 18 microsatellite loci data set for the prion taxa (Pachyptila) in this study. Genetic structure was modelled in STRUCTURE assuming admixture and the partitioning of genetic diversity into two (K = 2) and three (K = 3) populations (K4–10 shown in Supplemental Fig. S1). Each line on the plot represents an individual multilocus profile
We further tested the structure of populations by analysing the molecular variance in our microsatellite data set in an AMOVA framework. Among the various grouping scenarios tested, the structure separating P. macgillivrayi from St Paul Island from all other populations returned the highest among group variance (Fct). This value was only slightly lower when Gough medium-billed prions were included in the same group as P. macgillivrayi (Table 3). Including Gough P. vittata into this group returned a much lower Fct value. These results are further supported by pairwise FST values among populations, which were lowest between P. macgillivrayi and Gough medium-billed prions, higher between P. macgillivrayi and Gough P. vittata and highest in comparisons to the other P. vittata populations (Table 4).
Using BAYESASS, we estimated bidirectional interspecific migration rates, which were generally low, except in the case of the Gough medium-billed prions, where up to 28% of that gene pool could have been derived through unidirectional gene flow from P. macgillivrayi from St Paul, and up to 8% derived from Gough P. vittata (Table 5; results for all studied populations are provided in the Supplemental Material, Table S4).
The most likely bill width in the common ancestor to all extant Pachyptila species was 14.2 mm (median posterior ancestral bill width estimated by BAYESTRAITS; Q1 = 11.9, Q3 = 16.4; Fig. 5). Including Halobaena caerulea, an even narrower ancestral median bill width of 12.8 mm was inferred (Q1 = 8.7, Q3 = 17; Supplemental Material, Fig. S2).
Bill colour varies considerably among prions at Gough and Tristan (Supplemental Material, Fig. S3). Although many P. vittata have blackish bills (Fig. S3A, B) similar to this taxon in New Zealand (Marchant and Higgins 1990; Shirihai 2007, but see Supplemental Material, Fig. S4), some have bluish-grey sides to the bill, even among birds breeding in spring at Inaccessible Island, Tristan da Cunha (Fig. S3C), and one unusual bird has been photographed on Gough with bright blue sides to the bill (Fig. S3D). Gough medium-billed birds seemingly show less variation, with generally lead-grey bills with variably paler blue-grey bill sides and tips (Fig. S3E, F). However, blackish bills are also not confined to P. vittata, as some fairly small-billed birds (likely P. desolata) photographed at sea near the Crozets in December 2016 had blackish bills (Supplemental Material, Fig. S5A, B) compared to typical P. desolata (Fig. S5C, D). Consequently, bill colour was not a reliable trait for distinguishing these taxa.
Discussion
Are Gough Island medium-billed prions an undiscovered population of P. macgillivrayi?
The medium-billed prions from Gough Island clustered in a monophyletic clade, which was sister to P. macgillivrayi (Fig. 3), and microsatellite genotypes from 10 Gough medium-billed individuals consistently clustered them with P. macgillivrayi at K ≥ 3 (Fig. 4). These results suggest that the Gough medium-billed prions belong to the same evolutionary lineage as P. macgillivrayi with which it shared a common ancestor between 0.8 and 3.3 Mya. The use of only mtDNA and a single fossil calibration could potentially also have led to biases in divergence times, and although the relative branching patterns are independent of divergence time, we suggest the reader consider the full 95% HPD when interpreting the timing of evolutionary events. Despite this potentially old divergence, microsatellite variation among Gough medium-billed prions did not differ from that of P. macgillivrayi. This suggests that Gough medium-billed prions represent a hitherto undiscovered population of MacGillivray’s prion. These results also imply that the P. macgillivrayi medium-billed phenotype must have evolved prior to the Pleistocene divergence of the Gough medium-billed population.
Colonisation of Gough Island by P. macgillivrayi
Our analysis of gene flow among P. macgillivrayi, Gough medium-billed prions and P. vittata confirmed admixture analyses in STRUCTURE that indicated low levels of gene flow into the P. macgillivrayi population on St Paul Island. However, the same analysis revealed that a significant proportion of the Gough Island medium-billed gene pool stemmed from St Paul P. macgillivrayi. This could represent ongoing gene flow between these two populations, or it could be the molecular signature of the original colonisation event that brought medium-billed prions to Gough Island. However, despite Gough medium-billed population being much larger (875,000 breeding pairs; Caravaggi et al. 2019) than the P. macgillivrayi population on St Paul (a few hundred birds; Tollu 1984; Shirihai 2007), the inferred migration into the Gough medium-billed population was unidirectional, which is more compatible with a colonisation event as ongoing gene flow is more likely to result in bi-directional gene flow. On the other hand, we also detected lower levels of unidirectional gene flow from Gough P. vittata into the Gough medium-billed population (Table 5), which is supported by their low pairwise FST value of 0.038 (Table 4). This incoming gene flow from P. vittata may also explain the overlap in bill widths among the smallest P. vittata and the largest Gough medium-billed prions (Fig. 2). Because these populations are sympatric, breeding on the same island (Ryan et al. 2014; Jones et al. 2020), this result is likely to represent ongoing gene flow, with directionality determined by assortative mating of medium-billed females with natal philopatry of hybrid offspring. Larger sample sizes and a modelling approach similar to that of Masello et al. (2019) could help distinguish these scenarios.
Medium-billed Pachyptila species evolved independently through convergence
Bayesian trait reconstruction suggested an ancestral maternal bill width for Pachyptila of around 14 mm, which is roughly at a transition between extant narrow (11 mm) and medium-billed (14.2–18 mm) species (Fig. 2, Supplemental Material, Table S2). However, considering its wide range (Q1 = 11.9, Q3 = 16.4; Fig. 5), this ancestral bill width (14 mm) is still narrower than that of extant P. macgillivrayi (18 mm) and P. salvini (16.7 mm; Fig. 2, Supplemental Material, Table S2). Further ancestral trait reconstruction including the outgroup taxon suggests that bill width was even narrower further back in time, strongly implying that extant Pachyptila were derived from a thinner-billed ancestor, with medium bills > 16 mm and broad bills > 21 mm being derived character states. This is consistent with the evolution of specialised filtering lamellae along the sides of the upper mandible among the broader-billed Pachyptila taxa as a derived trait. However, we caution that our ancestral state reconstruction was based on a posterior distribution of mtDNA data, representing only maternal evolutionary events. A species tree reconstructed from autosomal loci would better account for introgression and so may be different to the mtDNA tree presented here. Whole genome sequencing of all species within the genus, with the aim of reconstructing a reliable species tree is thus a research priority.
Both mtDNA and nuclear markers revealed an evolutionary history for P. macgillivrayi that is distinct from that of P. salvini, despite the two species possessing similar bill phenotypes. Maternally, P. macgillivrayi is sister to P. vittata, whereas P. salvini is sister to P. desolata (Fig. 3). Microsatellite variation partitions P. macgillivrayi and Gough medium-billed as distinct from other species at K = 3, and with very little incoming gene flow, unlike the multilocus profile of P. salvini, where higher admixture can be observed (Fig. 4). Harper (1980) was thus correct in placing P. macgillivrayi as closer to P. vittata rather than P. salvini (Roux et al. 1986). Coalescent simulations of microsatellite DNA showed that P. salvini’s medium bill width (mean: this study, 16.7 mm, Fig. 2, 17.1 mm in Masello et al. (2019)) evolved through hybridisation of narrower- and broader-billed species, presumably conferring a feeding advantage as its intermediate bill width allowed it to feed on a wider variety of prey species than either of its parent species (Masello et al. 2019). Thus unlike P. salvini, which has a hybrid origin, our results show that P. macgillivrayi evolved after divergence from thinner-billed species (Fig. 3). Given this evolutionary scenario, it is likely that the broad bill of P. vittata was derived from a medium-billed ancestor, possibly similar in bill width to P. macgillivrayi. Therefore, the positions of similarly medium-billed P. salvini and P. macgillivrayi in different clades of the Pachyptila phylogeny strongly suggest mutually exclusive evolutionary histories for these species and, thus, the independent evolution of their phenotypes. As with P. salvini, an intermediate bill width (18 mm; Fig. 2, Supplemental Material, Table S2) might allow P. macgillivrayi to feed on a similarly wide range of prey species, providing a selective driver for the convergent evolution of the medium-billed phenotype.
Status and conservation P. macgillivrayi
The conservation status of P. macgillivrayi was only assessed for the IUCN Red List of Threatened Species in 2016, because prior to the findings of our study it was not recognised as a species by BirdLife International. The population that breeds in the Indian Ocean is small; about 1000 pairs that were initially confined (150 pairs) to La Roche Quille, a rock stack 150 m off the coast of St Paul Island (Tollu 1984; Jiguet et al. 2007; Barbraud et al. 2021). The species was once abundant (> 105 pairs) on Amsterdam Island and St Paul, but was extirpated from both islands by introduced predators (Worthy and Jouventin 1999; Jiguet et al. 2007). A significant number of prions have recolonised St Paul from La Roche Quille following the eradication of Black Rats Rattus rattus from St Paul in 1997 and European Rabbits Oryctolagus cuniculus in 1999 (Micol and Jouventin 2002; Griffiths 2011; Barbraud et al. 2021). Unfortunately, house mice Mus musculus remain on St Paul Island (Micol and Jouventin 2002), and predation by mice may slow or even halt the recovery of prions (Dilley et al. 2015). The population also is at risk from vagrant falcons that occasionally reach the island (Jiguet et al. 2007; Barbraud et al. 2021).
The population size of P. macgillivrayi on Gough Island is not well known, but based on their proportions in skua prey remains, they comprise at least 20% of prions breeding on the island (Jones 2018). Cuthbert (2004) estimated there were 1.75 million pairs of prions breeding on Gough Island in 2000. This estimate might be somewhat inflated, and numbers of prions probably are decreasing due to heavy predation on their eggs and chicks by introduced house mice (Cuthbert et al. 2013; Dilley et al. 2015), but it is still likely that Gough supports at least 105 pairs of P. macgillivrayi; Caravaggi et al. (2019) estimated 875,000 pairs. Gough Island is thus home to more than 99% of the species’ global population. MacGillivray’s prion qualifies as Endangered under the IUCN Red List of Threatened Species Criterion B2a (two populations with a total breeding area < 70 km2). The species has experienced very poor breeding success (0–15%) in recent years due to mouse predation on Gough Island (Dilley et al. 2015), so its population is almost certainly decreasing. If so, it also qualifies as Endangered under Criterion B2b. The recognition of yet another globally threatened species that is virtually confined to Gough Island provides further impetus for the need to eradicate mice from the island (Jones et al. 2021).
Conclusions
Our results suggest a different evolutionary history for P. macgillivrayi and P. salvini. We show that the intermediate bill width of P. macgillivrayi is not the product of interspecies introgression or hybrid speciation, but evolved through divergence. Remarkably, the newly described Gough Island population of medium-billed prions belong to the same evolutionary lineage as P. macgillivrayi, representing a new population of MacGillivray’s prion that originated through a colonisation event from St Paul Island. Unidirectional gene flow from Gough P. vittata into the Gough medium-billed population exists and merits future investigation. Given that the relict P. macgillivrayi population in the Indian Ocean is very small, our results demonstrating that the newly discovered medium-billed prions population on Gough corresponds to a new population of MacGillivray’s prion are of utmost relevance for the conservation of this species. Our results provide further evidence for the need to eradicate introduced house mice from Gough Island (Holmes et al. 2019; Jones et al. 2021).
Availability of data and material
DNA sequences: GenBank accession numbers are provided in Table 1. All data are available in the manuscript or in the Supplementary information file.
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Barbraud C, Delord K, Le Bouard F, Harivel R, Demay J, Chaigne A, Micol T (2021) Seabird population changes following mammal eradication at oceanic Saint-Paul Island, Indian Ocean. J Nat Conserv 63:126049
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2004) Laboratoire Génome, populations, interactions, CNRS UMR 5171. Université de Montpellier II, Montpellier
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D et al (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLOS Comp Biol 10:e1003537
Bretagnolle V, Zotier R, Jouventin P (1990) Comparative population biology of four prions (Genus Pachyptila) from the Indian Ocean and consequences for their taxonomic status. Auk 107:305–316
Burg T, Croxall J (2001) Global relationships amongst black-browed and grey-headed albatrosses: analysis of population structure using mitochondrial DNA and microsatellites. Mol Ecol 10:2647–2660
Caravaggi A, Cuthbert RJ, Ryan PG, Cooper J, Bond AL (2019) The impacts of introduced house mice on the breeding success of nesting seabirds on Gough Island. Ibis 161:648–661
Cherel Y, Bocher P, De Broyer C, Hobson KA (2002) Food and feeding ecology of the sympatric thin-billed Pachyptila belcheri and Antarctic P. desolata prions at Iles Kerguelen, Southern Indian Ocean. Mar Ecol Progr Ser 228:263–281
Cox J (1980) Some remarks on the breeding distribution and taxonomy of the prions (Procellariidae: Pachyptila). Rec S Aust Mus 18:91–121
Cuthbert R (2004) Breeding biology of the Atlantic Petrel, Pterodroma incerta, and a population estimate of this and other burrowing petrels on Gough Island, South Atlantic Ocean. Emu 104:221–228
Cuthbert RJ, Louw H, Lurling J, Parker G, Rexer-Huber K, Sommer E et al (2013) Low burrow occupancy and breeding success of burrowing petrels at Gough Island: a consequence of mouse predation. Bird Conserv Int 23:113–124
Dilley BJ, Davies D, Bond AL, Ryan PG (2015) Effects of mouse predation on burrowing petrel chicks at Gough Island. Antarct Sci 27:543–553
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Gen Res 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Falla RA (1940) The genus Pachyptila Illiger. Emu 40:218–236
Fleming CA (1941) The phylogeny of the prions. Emu 41:134–155
Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Fullagar PJ (1972) Identification of prions—Pachyptila spp. Aust Bird Bander 10:36–39
Gartshore N, Steele W, Klages N (1988) Summer diet of the Salvin’s prion at sub-Antarctic Marion Island. S Afr J Zool 23:309–313
Genovart M, Thibault J-C, Igual JM, Bauzà-Ribot MdM, Rabouam C, Bretagnolle V (2013) Population structure and dispersal patterns within and between atlantic and mediterranean populations of a large-range pelagic seabird. PLoS ONE 8:e70711
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Griffiths R (2011) Targeting multiple species—a more efficient approach to pest eradication. Island invasives: eradication and management. International Union for Conservation of Nature, Gland, pp 172–176
Harper PC (1980) The field identification and distribution of the prions (genus Pachyptila), with particular reference to the identification of storm-cast material. Notornis 27:235–286
Holmes ND, Spatz DR, Oppel S, Tershy B, Croll DA, Keitt B et al (2019) Globally important islands where eradicating invasive mammals will benefit highly threatened vertebrates. PLoS ONE 14:e0212128
Howell SNG, Zufelt K (2019) Oceanic birds of the world: a photo guide. Princeton University Press, Princeton
Imber M (1981) Diets of stormpetrels Pelagodroma and Garrodia and of prions Pachyptila (Procellariiformes). Proc Symp Bird Sea Shore 1:63–88
Inchausti P, Weimerskirch H (2002) Dispersal and metapopulation dynamics of an oceanic seabird, the wandering albatross, and its consequences for its response to long-line fisheries. J Anim Ecol 71:765–770
Jiguet F, Robert A, Micol T, Barbraud C (2007) Quantifying stochastic and deterministic threats to island seabirds: last endemic prions face extinction from falcon peregrinations. Anim Conserv 10:245–253
Jones CW, Risi MM, Osborne AM, Ryan PG, Oppel S (2021) Mouse eradication is required to prevent local extinction of an endangered seabird on an oceanic island. Anim Conserv 24:637–645
Jones CW, Phillips RA, Grecian WJ, Ryan PG (2020) Ecological segregation of two superabundant, morphologically similar, sister seabird taxa breeding in sympatry. Mar Biol 167:e45
Jones CWP (2018) Comparative ecology of Pachyptila species breeding sympatrically at Gough Island. MSc thesis, University of Cape Town
Klages NTW, Cooper J (1992) Bill morphology and diet of a filter-feeding seabird: the broad-billed prion Pachyptila vittata at South Atlantic Gough Island. J Zool 227:385–396
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15:1179–1191
Kunte K, Shea C, Aardema ML, Scriber JM, Juenger TE, Gilbert LE et al (2011) Sex chromosome mosaicism and hybrid speciation among Tiger Swallowtail butterflies. PLOS Genet 7:e1002274
Lamichhaney S, Han F, Webster MT, Andersson L, Grant BR, Grant PR (2018) Rapid hybrid speciation in Darwin’s finches. Science 359:224–228
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Marchant S, Higgins PJ (1990) Volume 1, ratites to ducks; part A, ratites to petrels. In: Marchant S, Higgins PJ (eds) Handbook of Australian, New Zealand and Antarctic Birds. Oxford University Press, Melbourne
Masello JF, Quillfeldt P, Sandoval-Castellanos E, Alderman R, Calderón L, Cherel Y et al (2019) Additive traits lead to feeding advantage and reproductive isolation, promoting homoploid hybrid speciation. Mol Biol Evol 36:1671–1685
Mathews GM (1934) A check-list of the order Procellariiformes. Nov Zool 39:153–206
Mayr E (1942) Systematics and the origin of species, from the viewpoint of a zoologist. Columbia University Press, New York
Mayr E (1963) Animal species and evolution. Harvard University Press, Cambridge
Micol T, Jouventin P (2002) Eradication of rats and rabbits from Saint-Paul Island, French Southern territories. In: Veitch CR, Clout MN (eds) Turning the tide: the eradication of invasive species. IUCN SSC Invasive Species Specialist Group, Gland, pp 199–205
Moodley Y, Masello JF, Cole TL, Calderon L, Munimanda GK, Thali MR et al (2015) Evolutionary factors affecting the cross-species utility of newly developed microsatellite markers in seabirds. Mol Ecol Res 15:1046–1058
Morgan WL, Ritz DA (1982) Comparison of the feeding apparatus in the muttonbird, Puffinus tenuirostris (Temminck) and the fairy prion, Pachyptila turtur (Kuhl) in relation to the capture of the krill, Nyctiphanes australis Sars. J Exp Mar Biol Ecol 59:61–75
Murphy RC (1936) Oceanic Birds of South America, vol 2. Macmillan, New york
Nadeau NJ, Whibley A, Jones RT, Davey JW, Dasmahapatra KK, Baxter SW et al (2012) Genomic islands of divergence in hybridizing Heliconius butterflies identified by large-scale targeted sequencing. Phil Trans R Soc B Biol Sci 367:343–353
Nolte AW, Freyhof J, Tautz D (2006) When invaders meet locally adapted types: rapid moulding of hybrid zones between sculpins (Cottus, Pisces) in the Rhine system. Mol Ecol 15:1983–1993
Olson SL (1983) Fossil seabirds and changing marine environments in the Late Tertiary of South Africa. S Afr J Sci 79:399–402
Olson SL (1985a) An early Pliocene marine avifauna from Duinefontein, Cape Province, South Africa. Ann S Afr Mus 95:147–164
Olson SL (1985b) Early Pliocene Procellariiformes (Aves) from Langebaanweg, South-western Cape Province, South Africa. Ann S Afr Mus 95:123–145
Pagel M (1999) Inferring the historical patterns of biological evolution. Nature 401:877–884
Pagel M, Meade A, Barker D (2004) Bayesian estimation of ancestral character states on phylogenies. Syst Biol 53:673–684
Patel S, Waugh J, Millar CD, Lambert DM (2010) Conserved primers for DNA barcoding historical and modern samples from New Zealand and Antarctic birds. Mol Ecol Resour 10(3):431–438
Penhallurick J, Wink M (2004) Analysis of the taxonomy and nomenclature of the Procellariiformes based on complete nucleotide sequences of the mitochondrial cytochrome b gene. Emu 104:125–147
Petit RJ, Excoffier L (2009) Gene flow and species delimitation. Trends Ecol Evol 24:386–393
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Quillfeldt P, Cherel Y, Delord K, Weimerkirch H (2015) Cool, cold or colder? Spatial segregation of prions and blue petrels is explained by differences in preferred sea surface temperatures. Biol Lett 11:20141090
Quillfeldt P, Moodley Y, Weimerskirch H, Cherel Y, Delord K, Phillips R et al (2017) Does the genetic structure reflect differences in non-breeding movement? A case study in small, highly mobile seabirds. BMC Evol Biol 17:e160
Quillfeldt P, Weimerskirch H, Delord K, Cherel Y (2020) Niche switching and leapfrog foraging: movement ecology of sympatric petrels during the early breeding season. Mov Ecol 8:e23
R Development Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67(5):901
Rheindt FE, Austin JJ (2005) Major analytical and conceptual shortcomings in a recent taxonomic revision of the Procellariiformes—a reply to Penhallurick and Wink (2004). Emu 105:181–186
Ridoux V (1994) The diets and dietary segregation of seabirds at the Subantarctic Crozet Islands. Mar Ornithol 22:1–192
Rieseberg LH, Church SA, Morjan CL (2004) Integration of populations and differentiation of species. New Phytol 161:59–69
Rosenzweig ML (2001) Loss of speciation rate will impoverish future diversity. P Natl Acad Sci USA 98:5404–5410
Roux J-P, Mougin J-L, Bartle J (1986) Le prion de MacGillivray. Données Taxinomiques L’Oiseau et RFO 56:379–383
Ryan P, Bourgeois K, Dromzée S, Dilley B (2014) The occurrence of two bill morphs of prions Pachyptila vittata on Gough Island. Polar Biol 37:727–735
Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA et al (2014) Genomics and the origin of species. Nat Rev Gen 15:176–192
Shepherd LD, Lambert DM (2008) Ancient DNA and conservation: lessons from the endangered kiwi of New Zealand. Mol Ecol 17(9):2174–2184
Shirihai H (2007) A complete guide to Antarctic wildlife, 2nd edn. A&C Black, London
Szpiech ZA, Jakobsson M, Rosenberg NA (2008) ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics 24:2498–2504
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tollu B (1984) La Quille (Île Saint Paul, Océan Indien), sanctuaire de populations relictes. L’oiseau Et RFO 54:79–85
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115
van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Warham J (1990) The petrels—their ecology and breeding systems. Academic Press, London
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Worthy TH, Jouventin P (1999) The fossil avifauna of Amsterdam Island, Indian Ocean. Smithson Contrib Paleob 89:39–65
Acknowledgements
We thank Thierry Micol (Terres Australes et Antarctiques Françaises) for helping with sample collection, Gopi K. Munimanda and Boglárka Bálint for assisting with laboratory work, and Ricardo Palma for useful discussions. Francesco Bonadonna and Charlene Gemard kindly shared bill width data of blue petrels obtained in the framework of the “French Polar Institute IPEV, project 354 ETHOTAAF” (to F. Bonadonna). PQ, JFM, TLC and LC were supported by the Deutsche Forschungsgemeinschaft (Germany), Heisenberg program (grant number DFG, Qu 148-5 to P.Q.). Logistical and financial support was obtained from the South African Department of Environmental Affairs, through the South African National Antarctic Programme. Permission to collect samples at Tristan and Gough Island was obtained from Tristan’s Conservation Department. All sampling met approved ethical standards.
Funding
Open Access funding enabled and organized by Projekt DEAL. PQ, JFM, TLC and LC were supported by the Deutsche Forschungsgemeinschaft (Germany), Heisenberg program (grant number DFG, Qu 148-5 to P.Q.). Logistical and financial support was obtained from the South African Department of Environmental Affairs, through the South African National Antarctic Programme. LDS was supported by a Rutherford Discovery Fellowship from the Royal Society of New Zealand.
Author information
Authors and Affiliations
Contributions
PQ, JFM, PR, AJDT and YM conceived and designed the study. PR, PQ, YC, RA, RC, BJD, MM, CMM, JN, RP, HW carried out the extensive fieldwork. JFM, LDS, LC and TLC and carried out the laboratory work. JFM, YM, and LDS carried out the bioinformatics analyses. YM, JFM and PR drafted the manuscript. All authors reviewed the final draft of the manuscript.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare that they have no competing interests.
Ethics approval
Permission to collect samples at Tristan and Gough Island was obtained from Tristan’s Conservation Department. All sampling met approved ethical standards.
Additional information
Communicated by Martine Collart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Masello, J.F., Ryan, P.G., Shepherd, L.D. et al. Independent evolution of intermediate bill widths in a seabird clade. Mol Genet Genomics 297, 183–198 (2022). https://doi.org/10.1007/s00438-021-01845-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01845-3