Abstract
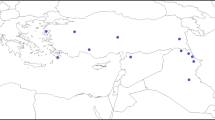
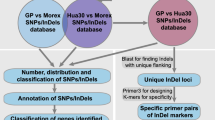
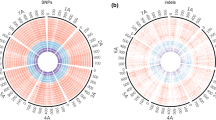
The wild species in the Triticeae tribe are tremendous resources for crop breeding due to their abundant natural variation. However, their huge and highly repetitive genomes have hindered the establishment of physical maps and the completeness of their genome sequences. To develop molecular markers for the efficient utilization of their valuable traits while avoiding their genome complexity, we assembled RNA sequences of ten representative accessions of Aegilops tauschii, the progenitor of the wheat D genome, and estimated single nucleotide polymorphisms (SNPs) and insertions/deletions (indels). The deduced unigenes were anchored to the chromosomes of Ae. tauschii and barley. The SNPs and indels in the anchored unigenes, covering entire chromosomes, were sufficient for linkage map construction, even in combinations between the genetically closest accessions. Interestingly, the resolution of SNP and indel distribution on barley chromosomes was slightly higher than on Ae. tauschii chromosomes. Since barley chromosomes are regarded as virtual chromosomes of Triticeae species, our strategy allows capture of genetic markers arranged on the chromosomes in order based on the conserved synteny. The resolution of these genetic markers will be comparable to that of the Ae. tauschii whose draft genome sequence is available. Our procedure should be applicable to marker development for Triticeae species, which have no draft sequences available.




Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178
Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: a web-based genome analysis tool for experimentalists. In: Current protocols in molecular biology , Chap 19.10, vol 89, pp 19.10.1–19.10.21
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510
Gaut BS (2002) Evolutionary dynamics of grass genomes. New Phytol 154:15–28
Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15:1451–1455
Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11:R86
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, Macmanes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Iehisa JCM, Shimizu A, Sato K, Nasuda S, Takumi S (2012) Discovery of high-confidence single nucleotide polymorphisms from large-scale de novo analysis of leaf transcripts of Aegilops tauschii, a wild wheat progenitor. DNA Res 19:487–497
Iehisa JCM, Shimizu A, Sato K, Nishijima R, Sakaguchi K, Matsuda R, Nasuda S, Takumi S (2014) Genome-wide marker development for the wheat D genome based on single nucleotide polymorphisms identified from transcripts in the wild wheat progenitor Aegilops tauschii. Theor Appl Genet 127:261–271
International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491:711–716
Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer KFX, Li D, Pan S, Zheng F, Hu Q, Xia X, Li J, Liang Q, Chen J, Wicker T, Gou C, Kuang H, He G, Luo Y, Keller B, Xia Q, Lu P, Wang J, Zou H, Zhang R, Xu J, Gao J, Middleton C, Quan Z, Liu G, Wang J, International Wheat Genome Sequencing Consortium, Yang H, Liu X, He Z, Mao L, Wang J (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496:91–95
Kersey PJ, Allen JE, Armean I, Boddu S, Bolt BJ, Carvalho-Silva D, Christensen M, Davis P, Falin LJ, Grabmueller C, Humphrey J, Kerhornou A, Khobova J, Aranganathan NK, Langridge N, Lowy E, McDowall MD, Maheswari U, Nuhn M, Ong CK, Overduin B, Paulini M, Pedro H, Perry E, Spudich G, Tapanari E, Walts B, Williams G, Tello-Ruiz M, Stein J, Wei S, Ware D, Bolser DM, Howe KL, Kulesha E, Lawson D, Maslen G, Staines DM (2015) Ensembl genomes 2016: more genomes, more complexity. Nucleic Acids Res 44:574–580
Kosugi S, Natsume S, Yoshida K, MacLean D, Cano L, Kamoun S, Terauchi R (2013) Coval: improving alignment quality and variant calling accuracy for next-generation sequencing data. PLoS One 8:e75402
Kumar S, Banks TW, Cloutier S (2012) SNP discovery through next-generation sequencing and its applications. Int J Plant Genomics. doi:10.1155/2012/831460
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup 1000 Genome Project Data Processing (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Luo M-C, Gu YQ, You FM, Deal KR, Ma Y, Hu Y, Huo N, Wang Y, Wang J, Chen S, Jorgensen CM, Zhang Y, McGuire PE, Pasternak S, Stein JC, Ware D, Kramer M, McCombie WR, Kianian SF, Martis MM, Mayer KFX, Sehgal SK, Li W, Gill BS, Bevan MW, Šimková H, Doležel J, Weining S, Lazo GR, Anderson OD, Dvorak J (2013) A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA 110:7940–7945
Matsumoto T, Tanaka T, Sakai H, Amano N, Kanamori H, Kurita K, Kikuta A, Kamiya K, Yamamoto M, Ikawa H, Fujii N, Hori K, Itoh T, Sato K (2011) Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol 156:20–28
Matsuoka Y, Nasuda S, Ashida Y, Nitta M, Tsujimoto H, Takumi S, Kawahara T (2013) Genetic basis for spontaneous hybrid genome doubling during allopolyploid speciation of common wheat shown by natural variation analyses of the paternal species. PLoS One 8:e68310
Matsuoka Y, Takumi S, Kawahara T (2015) Intraspecific lineage divergence and its association with reproductive trait change during species range expansion in central Eurasian wild wheat Aegilops tauschii Coss. (Poaceae). BMC Evol Biol 15:213
Mayer KFX, Martis M, Hedley PE, Šimková H, Liu H, Morris JA, Steuernagel B, Taudien S, Roessner S, Gundlach H, Kubaláková M, Suchánková P, Murat F, Felder M, Nussbaumer T, Graner A, Salse J, Endo T, Sakai H, Tanaka T, Itoh T, Sato K, Platzer M, Matsumoto T, Scholz U, Doležel J, Waugh R, Stein N (2011) Unlocking the barley genome by chromosomal and comparative genomics. Plant Cell 23:1249–1263
Mizuno N, Yamasaki M, Matsuoka Y, Kawahara T, Takumi S (2010) Population structure of wild wheat D-genome progenitor Aegilops tauschii Coss.: implications for intraspecific lineage diversification and evolution of common wheat. Mol Ecol 19:999–1013
Nishijima R, Iehisa JCM, Matsuoka Y, Takumi S (2014) The cuticular wax inhibitor locus Iw2 in wild diploid wheat Aegilops tauschii: phenotypic survey, genetic analysis, and implications for the evolution of common wheat. BMC Plant Biol 14:246
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842
Sehgal D, Vikram P, Sansaloni CP, Ortiz C, Saint Pierre C, Payne T, Ellis M, Amri A, Petroli CD, Wenzl P, Singh S (2015) Exploring and mobilizing the gene bank biodiversity for wheat improvement. PLoS One 10:e0132112
Takagi H, Uemura A, Yaegashi H, Tamiru M, Abe A, Mitsuoka C, Utsushi H, Natsume S, Kanzaki H, Matsumura H, Saitoh H, Cano LM, Kamoun S, Terauchi R (2013) Methods MutMap-Gap: whole-genome resequencing of mutant F2 progeny bulk combined with de novo assembly of gap regions identifies the rice blast resistance gene Pii. New Phytol 200:276–283
Takumi S, Koyama K, Fujiwara K, Kobayashi F (2011) Identification of a large deletion in the first intron of the Vrn-D1 locus, associated with loss of vernalization requirement in wild wheat progenitor Aegilops tauschii Coss. Genes Genet Syst 86:183–195
Tsunewaki K (1966) Comparative gene analysis of common wheat and its ancestral species. II. Waxiness, growth habit and awnedness. Jpn J Bot 19:175–229
Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR (2015) BrAD-seq: Breath Adapter Directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front Plant Sci 6:366
Wicker T, Mayer KFX, Gundlach H, Martis M, Steuernagel B, Scholz U, Šimková H, Kubaláková M, Choulet F, Taudien S, Platzer M, Feuillet C, Fahima T, Budak H, Dolezel J, Keller B, Stein N (2011) Frequent gene movement and pseudogene evolution is common to the large and complex genomes of wheat, barley, and their relatives. Plant Cell 23:1706–1718
Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21:1859–1875
Yang C, Zhao L, Zhang H, Yang Z, Wang H, Wen S, Zhang C, Rustgi S, von Wettstein D, Liu B (2014) Evolution of physiological responses to salt stress in hexaploid wheat. Proc Natl Acad Sci 111:11882–11887
Zhang J, Kobert K, Flouri T, Stamatakis A (2014) PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620
Acknowledgments
Computations for the RNA sequencing assembly and alignments of short reads were performed on the NIG supercomputer at the ROIS National Institute of Genetics.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Funding
This work was supported by a Grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan [Grant-in-Aid for Scientific Research (B) Nos. 25292008 and 16H04862] to ST, and by MEXT as part of a Joint Research Program implemented at the Institute of Plant Science and Resources, Okayama University, Japan.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nishijima, R., Yoshida, K., Motoi, Y. et al. Genome-wide identification of novel genetic markers from RNA sequencing assembly of diverse Aegilops tauschii accessions. Mol Genet Genomics 291, 1681–1694 (2016). https://doi.org/10.1007/s00438-016-1211-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-016-1211-2