Abstract
Different species of amoebae belonging to the genus Acanthamoeba are widely distributed in many parts of the world and known as free-living organisms. Some strains of the protozoans may exist as parasites and cause risk to human health as causative agents of serious human diseases. Currently, in Poland, there is no sufficient information about the distribution of Acanthamoeba strains and their genotypes in the environment. Therefore, 20 environmental surface water samples were collected from different sites located at five water reservoirs in Gdynia, Sopot, and Gdańsk (northern Poland). The material was cultured to obtain Acanthamoeba isolates that were then specifically analyzed with both PCR and real-time PCR assays. Of the 20 samples examined, Acanthamoeba DNA was found in 13 samples tested with the use of real-time PCR; in 10 of them, DNA of the amoeba was also detected using PCR technique. The comparison with sequences available in the GenBank confirmed that the PCR products are fragments of Acanthamoeba 18S rRNA gene and that isolates represent T4 genotype, known as the most common strains related to AK cases. This is the first investigation in Poland describing Acanthamoeba detection in environmental water samples with molecular techniques and genotyping. The results indicate that surface water in Poland may be a source of acanthamoebic strains potentially pathogenic for humans.
Similar content being viewed by others
Introduction
Different species of the genus Acanthamoeba exist as free-living protists that are widely prevalent in natural and man-made environments. The amoebae were detected in soil, water, animals, fruits and vegetable, air, dust, hospital environment, contact lens solutions, and eyewash stations as well as isolated from human body surfaces and various human tissues and cavities (Larkin et al. 1990; Gray et al. 1995; Van Hamme et al. 2001; Chomicz et al. 2002; Schuster and Visvesvara 2004; Lorenzo-Morales et al. 2005b; Khan 2006; Carlesso et al. 2010; Costa et al. 2010; Stockman et al. 2011; Kao et al. 2012). The Acanthamoeba genus involves species that complete their life cycles in the outer environment. However, in predisposing circumstances, some strains of Acanthamoeba may be able to enter the human body from environmental sources and exist as parasites.
The high rate of specific antibodies (50–100 %) in healthy populations confirmed that humans are frequently exposed to Acanthamoeba (Alizadeh et al. 2001; Chappell et al. 2001; Brindley et al. 2009). Physiological and biochemical investigations carried out to determine pathogenic Acanthamoeba strains included assessment of temperature and osmo-tolerance as well as the secretion of proteolytic enzymes (Khan et al. 2000; Clarke and Niederkorn 2006; Chomicz et al. 2010). In immunocompetent individuals, infections with these amoebae are commonly asymptomatic and self-limited. However, some strains of Acanthamoeba species play a role in public health as causative agents of serious human diseases: the severe, usually fatal granulomatous amoebic encephalitis (GAE) recorded among immunocompromised patients and Acanthamoebic keratitis (AK), the non-opportunistic, vision-threatening corneal infection. The latter is found particularly in contact lens wearers, which make up 95 % of cases. Due to the popularity of contact lenses, that is considered an important risk factor in contracting corneal disease, incidents of an acute AK are increasingly recognized in various parts of the world, including Poland (Marciano-Cabral and Cabral 2003; Schuster and Visvesvara 2004; Wesołowska et al. 2006; Ibrahim et al. 2007; Visvesvara and Schuster 2008; Chomicz et al. 2012; Mahmoudi et al. 2012; Szaflik et al. 2012). Furthermore, the amoebae may transmit some yeast, viruses, and bacteria strains pathogenic for humans as well as oocysts of protozoan parasite Cryptosporidium; some of these microorganisms are able to not only survive but even proliferate within these amoebae (Gómez-Couso et al. 2007; Khan 2009; Scheid and Schwarzenberger 2011; Anacarso et al. 2012).
For years, an increasing number of isolates of Acanthamoeba species have been recognized and classified using morphological criteria, mainly cyst size and the number of arms within a single cyst. Amoebae belonging to the Acanthamoeba genus were placed into three morphological groups I, II, III (Pussard and Pons 1977; Page 1988; Khan 2009). Recently, this way of Acanthamoeba classification has begun to be seen as unreliable due to, among others, subtle differences in cyst features; non-morphological procedures were also used, including biochemical, immunological, and physiological criteria being inconclusive in the identification of Acanthamoeba species (Stratford and Griffiths 1978; Marciano-Cabral and Cabral 2003; Walochnik et al. 2002; Chan et al. 2010). The situation is changing with the recent development of specific and sensitive methods for classification of Acanthamoeba isolates at the molecular level. The new approach is based on genotype associations. Many PCR assays were designed to detect Acanthamoeba isolates in clinical and environmental samples with the high specificity and efficiency at the genus level (Vodkin et al. 1992; Howe et al. 1997; Pélandakis et al. 2000; Schroeder et al. 2001; Khan and Paget 2002; Khan et al. 2002; Pélandakis and Pernin 2002; Booton et al. 2005; Qvarnstrom et al. 2006). Also, real-time PCR was developed as a fast tool for differential identification of free-living amoebae (Qvarnstrom et al. 2006; Rivière et al. 2006). Recently, the most frequently used technique for the characterization Acanthamoeba isolates is phylogenic analysis based on the 18S rRNA gene sequence. The technique of molecular typing, used for the first time by Gast et al. (1996), indentified 17 genotypes (T1–T17) within the genus with a sequence divergence of >5 % (Stothard et al. 1998; Hewett et al. 2003; Khan 2009; Corsaro and Venditti 2010; Nuprasert et al. 2010). Of these, seven genotypes were found in patients with AK (T2, T3, T4, T5, T6, T11, T15) of which T4 was the highest frequency (Booton et al. 2005; Maghsood et al. 2005; Risler et al. 2013). At present, it is known that the majority of human diseases caused by infections with Acanthamoeba were associated with some isolates of the T4 genotype, i.e., more than 90 % of AK was linked with this genotype (Khan 2009). However, it is not clear why not all T4 isolates are human pathogens; among others, differences in virulence and in susceptibility to chemicals as well as complications associated with transmission by the amoebae potentially pathogenic microorganisms are investigated and discussed for this term (Maghsood et al. 2005; Khan 2009; Risler et al. 2013).
The number of clinical and environmental Acanthamoeba isolates is increasing as well as of studies concerning their prevalence in environmental samples using molecular techniques (Lorenzo-Morales et al. 2005a; Lorenzo-Morales et al. 2005b; Kawaguchi et al. 2009; Magliano et al. 2009; Niyyati et al. 2009; Chan et al. 2010; Rahdar et al. 2012). In Poland, only a few investigations regarding the occurrence of Acanthamoeba in environmental samples have been undertaken, based on the morphological criteria of identification of the amoebae and bioassays (Kasprzak and Mazur 1972; Befinger et al. 1986). No molecular techniques were used. Therefore, it is important to carry out such examinations in order to detect the presence of amoebae in the environment and identify their species and genotype.
The aim of our study was to estimate the occurrence of the Acanthamoeba spp. in chosen natural water reservoirs in Poland using molecular techniques. The investigations were also carried out to determine genotypes of the obtained environmental isolates in terms of their association with those genotypes indicating potential threat to human health.
Material and methods
Laboratory isolates
Different laboratory strains of free-living amoebae were used in the study. DNA isolated from these strains served as controls in molecular investigations. Four Acanthamoeba laboratory isolates Acanthamoeba castellanii Neff, Acanthamoeba polyphaga-98, Acanthamoeba rhysodes, and Acanthamoeba astronyxis-190 were cultivated axenically (Červa and Novak 1968) in the laboratory of the Department of Tropical Parasitology, Medical University of Gdansk. Isolates of Naegleria: N. fowleri (strains: V212, V004, V414, V087), Naegleria italica, Naegleria lovaniensis, and Naegleria dunnebackei as well as Balamuthia mandrillaris were obtained from the Centers for Disease Control and Prevention (CDC, Atlanta, GA, USA).
Sampling and isolation of Acanthamoeba from the environmental material
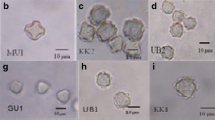
Twenty surface water samples were collected in June and July 2006. Environmental samples of water with sediment in proportion 1:1 were taken from the Kacza River (n = 5), Morskie Oko Pond (n = 5), Wysockie Lake (n = 2), Jasień Lake (n = 3), and Głębockie Lake (n = 5) located in the area of three cities: Gdynia, Sopot, and Gdańsk (northern Poland). The samples were taken from two different sites at each location (Table 1). The material was placed in 100-ml sterile polypropylene bottles and transported to the laboratory. Each sample was deposited onto two non-nutrient agar plates seeded with Aerobacter aerogenes. The material was placed along the diameter of the plate as a 0.5-cm band. Next, one plate was incubated at room temperature and the second at 37 °C. The plates were monitored daily under a light microscope for Acanthamoeba growth. Then, agar blocks containing Acanthamoeba were cut out and inoculated on new non-nutrient agar plates seeded with A. aerogenes until a large number of amoebae were observed. Next, they were washed out with distilled water and the aliquots kept at 4 °C for further examinations.
Molecular detection of Acanthamoeba
DNA extraction
DNA extraction was performed using the commercial Genomic Mini Kit (A&A Biotechnology, Gdynia, Poland) according to the manufacturer’s instructions. Then, DNA was stored at −20 °C.
DNA amplification
For specific detection of Acanthamoeba DNA, two non-commercial methods PCR and real-time PCR developed by Schroeder et al. (2001) and Qvarnstrom et al. (2006), respectively, were used in this study as described below.
PCR reaction was undertaken with the use of the primers JDP1 (5′GGCCCAGATCGTTT ACCGTGAA3′) and JDP2 (5′TCTCACAAGCTGCTAGGGAGTCA3′) targeting ~450 bp fragment of Acanthamoeba 18S rRNA gene (Schroeder et al. 2001). The amplification reaction mixture consisted of 2.5 μl of 10×PCR buffer for RUN polymerase (A&A Biotechnology, Poland), 0.25 mM of each dNTP (Fermentas, Lithuania), 0.2 μM of each primer (Metabion, Germany) 0.75U of RUN polymerase (A&A Biotechnology, Poland), and 2 μl of template DNA in a 25-μl reaction volume. Amplifications were performed according to the original previously described conditions in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, USA). PCR products were analyzed using GelDoc-It Imaging Systems (UVP, USA) after electrophoresis on agarose gel (Sigma, St. Louis, Missouri) stained with ethidium bromide.
Real-time PCR was performed with the use of pair of primers AcantF900 (5′CCCAGATCGTTTACCGTGAA3′) and AcantR1100 (5′TAAATATTAATGCCCC CAACTATCC3′) targeting 180-bp fragment of the 18S rRNA gene and the fluorescently labeled TaqMan probe (5′Cy5 CTGCCACCGAATACATTAGCATGG BHQ33′) (Qvarnstrom et al. 2006). The amplification reaction mixture consisted of 12.5 μl of 2× Brilliant II QPCR Master Mix (Startagene, USA), 400 nM of each primer (Metabion, Germany), 80 nM of TaqMan probe (Metabion, Germany), and 2 μl of template DNA in a 25-μl reaction volume. Amplification was performed with an initial polymerase activation step (10 min at 95 °C), followed by 40 cycles of denaturation (15 s at 95 °C), and hybridization/extension (1 min at 63 °C) in an Mx3005P thermocycler (Stratagene, USA). PCR products were analyzed using MxPro QPCR Software. The cycle threshold (CT) value, determining the cycle number at which the reporter’s fluorescence exceeds the threshold value, was recorded. A sample was considered positive if the CT value was <40.
All PCR/real-time PCR experiments were performed with the inclusion of Acanthamoeba positive controls to ensure correct functionality of the reaction, Naegleria- and Balamuthia-positive controls to check specificity of the method as well as negative controls to ensure no contamination of the PCR components.
Sequencing
Direct sequencing was performed for eight obtained isolates. Before sequencing, PCR products were cleaned with the Cleanup Kit protocol (A&A Biotechnology, Poland). Cycle sequencing was performed with the use of the amplification primers and BigDye v.3.0 Terminator Cycle Sequencing Kit (Applied Biosystems, USA) in the GeneAmp PCR System 9700 thermocycler (Applied Biosystems, USA), in both orientations. The products of the sequencing reaction were cleaned using an ExTerminator Kit (A&A Biotechnology), denatured, and subjected to analysis on an automatic ABI PRISM 310 DNA Sequencer (Applied Biosystems) using standard procedures as described by the manufacturer. The sequences obtained were then analyzed, aligned, and compared with data from GenBank using GeneStudio Pro Software (GeneStudio, Inc., Suwanee, Georgia).
Results
In 13 tested environmental samples, amoebae growth was observed at 37 °C, in six samples at room temperature, and in one sample, no amoebae growth was found (Table 1). The growth rate was graded by visual observation without enumeration of organisms.
The presence of Acanthamoeba DNA was recorded in 13 samples tested with the use of real-time PCR and in 10 samples using PCR. No product was found in DNA extracted from isolates of Naegleria and Balamuthia, which shows the specificity of chosen methods. The results confirmed that both methods chosen can be used to identify genus Acanthamoeba. However, real-time PCR was more sensitive than single-round PCR.
In order to determine the genotypes of Acanthamoeba isolates obtained from investigated environmental samples, sequencing of 10 PCR products was performed. In the case of the remaining three isolates (14, 15, and 19), that were positive only in real-time PCR, the amount of DNA was insufficient. The comparison of the obtained sequences with the Acanthamoeba sequences available in the GenBank confirmed that the detected PCR products were fragments of Acanthamoeba 18S rRNA gene. All sequenced isolates show 100 % homology with numbers of isolates belonging to the T4 genotype that is thought to be the cause most AK cases (Table 1). However, in the case of two isolates (samples 5 and 7), the sequences were shorter than in the other samples, and did not contain variable region (DF3 fragment). For this reason, it was impossible to exactly define their affiliation. Both of these isolates can be similar to each determined strain, or represent an additional one. Assuming the first option, four Acanthamoeba strains were defined. The variety of sequences was observed within one location, as well as between isolates obtained from different sampling sites (Table 1). It shows a high diversity within the Acanthamoeba genus.
Derived sequences containing DF3 fragment are deposited in GenBank under accession numbers KF924262–KF924269.
Discussion
To our knowledge, it is the first investigation in Poland describing Acanthamoeba detection in environmental samples using rapid molecular detection methods. The results of our findings confirmed that free-living amoebae of Acanthamoeba genus are present in surface water in Poland. Moreover, all sequenced strains belong to the T4 genotype of Acanthamoeba which is known as the most common genotype related to AK cases. It indicates that surface water in Poland may be a source of acanthamoebic infections in humans, and more samples should be tested to estimate the prevalence of this organism in this and other environmental matrices including air and soil. It is known that the classification of Acanthamoeba spp. based on morphological criteria is insufficient, while their identification is not problematic using different PCR assays, including real-time PCR. In this study, we used PCR assay with primers JDP, as well as real-time PCR, both based on the 18S rRNA gene fragments. Amplification was successful in all tested water samples using real-time PCR and in three less using PCR, which confirmed the suitability of the methods to estimate the incidence of Acanthamoeba in environmental material. However, from the point of view of higher sensitivity, timesaving, as well as additional control of genus specificity by using a molecular probe, real-time PCR has an advantage.
Nevertheless, since Gast et al. (1996) developed a classification scheme based on nuclear rRNA gene sequence, it has become the most frequently used technique for the characterization of Acanthamoeba isolates. Phylogenetic analysis based on 18S rRNA gene sequence has enabled the identification of 17 genotypes (T1–T17) within the genus with a sequence divergence of >5 %. In the present study, the investigated isolates of Acanthamoeba were genotyped by DNA sequencing with the use of JDP primers enclosing a fragment of the 18S rRNA gene. Primers JDP delimit ~450-bp fragment of the sequence containing diagnostics fragment 3 (DF3) which shows high variability within the genotypes of Acanthamoeba. A DF3 sequence was often used as a target in phylogenetic analysis in order to classify isolates to particular sequence types. However, some authors claim that this region is insufficient to discriminate isolates closely related T3, T4, and T11 genotypes (Schroeder et al. 2001; Risler et al. 2013). According to the authors, in some cases, Acanthamoeba isolates with the same DF3 sequence were not necessarily identical. For this reason, other variable regions of the 18S rRNA gene than DF3 should be used for correctly typing the isolates. This will be taken under consideration in our next study.
The majority of AK cases worldwide are connected with the genotype T4 and rarely with T2, T3, T6, and T11 (Walochnik et al. 2000; Khan et al. 2002; Maghsood et al. 2005; Ledee et al. 2009; Risler et al. 2013). It is not clear yet if the high isolation rate of the T4 genotype related to AK cases worldwide may be due to their greater prevalence in the environment, greater virulence or both (Khan 2006). However, many previous studies similarly showed that the T4 genotype is the most prevalent in the environment (Maghsood et al. 2005; Magliano et al. 2009; Niyyati et al. 2009; Rahdar et al. 2012).
Acanthamoeba spp. were found in different water samples worldwide. For instance, they were present in tap water in Brazil (Magliano et al. 2009); rivers, waterfall, and swimming pools in Iran (Maghsood et al. 2005; Niyyati et al. 2009; Rahdar et al. 2012); rivers and water treatment plants in Japan (Edagawa et al. 2009; Kawaguchi et al. 2009); and rivers, springs, wells, and water tanks in Nicaragua (Leiva et al. 2008). In this study, we investigated surface water samples taken from rivers and pond located at the area of three cities: Gdynia, Sopot, and Gdańsk in northern Poland. We found Acanthamoeba isolates in 65 % of samples tested. The results of sequencing the isolates showed 100 % similarity to sequences of isolates representing the T4 genotype deposited in GenBank. The differences noticed between the two locations (river in Gdynia and pond in Sopot) as well as within one location among two remote sampling sites confirm the heterogeneous nature of Acanthamoeba.
Human infections caused by Acanthamoeba remain poorly investigated in Poland. However, cases of acanthamoebic keratitis and studies with clinical isolates have been reported, and isolated strains were classified as T4 genotypes (Wesołowska et al. 2006; Szaflik et al. 2012). Environmental studies were rarely performed, mainly in the 1980s using morphological and physiological criteria. Among others, Befinger et al. (1986) found Naegleria and Acanthamoeba species in Lake Żarnowieckie and the Piaśnica River (northern Poland). Also, around Poznań (a city in western Poland), some lakes, a river and a canal used as recreational resorts, showed a common presence of Limax group amoebae (Kasprzak and Mazur 1972). Nowadays, the diversity and pathogenic potential of Acanthamoeba spp. and other free-living amoebae from Polish water reservoirs as well as other environmental matrices remain unknown.
This is the first report presenting results of our molecular studies on occurrence of Acanthamoeba genotypes in environment material. The results provide evidence that Acanthamoeba strains probably belonging to the T4 genotype, believed to be associated with AK cases, are present in the investigated water reservoirs, in northern Poland. For this reason, a larger number of environmental samples should be tested, taking into account different locations and water reservoir types, as well as studies relating to the diversity of Acanthamoeba strains and their prevalence and pathogenic potential.
References
Alizadeh H, Apte S, El-Agha MS, Li L, Hurt M, Howard K, Cavanagh HD, McCulley JP, Niederkorn JY (2001) Tear IgA and serum IgG antibodies against Acanthamoeba in patient with Acanthamoeba keratitis. Cornea 20:622–627
Anacarso I, de Niederhäusern S, Messi P, Guerrieri E, Iseppi R, Sabia C, Bondi M (2012) Acanthamoeba polyphaga, a potential environmental vector for the transmission of food-borne and opportunistic pathogens. J Basic Microbiol 52:261–268. doi:10.1002/jobm.201100097
Befinger M, Myjak P, Pietkiewicz H (1986) Occurrence of amphizoic amoebae in Lake Żarnowieckie. Bull Marit Trop Med 37:275–286
Booton GC, Visvesvara GS, Byers TJ, Kelly DJ, Fuerst PA (2005) Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J Clin Microbiol 43:1689–1693
Brindley N, Matin A, Khan NA (2009) Acanthamoeba castellanii: high antibody prevalence in racially and ethnically diverse populations. Exp Parasitol 121:254–256. doi:10.1016/j.exppara.2008.11.009
Carlesso AM, Artuso GL, Caumo K, Rott MB (2010) Potentially pathogenic acanthamoeba isolated from a hospital in Brazil. Curr Microbiol 60:185–190. doi:10.1007/s00284-009-9523-7
Červa L, Novak K (1968) Amoebic meningoencephalitis, sixteen fatalities. Science 160:92
Chan LL, Mak JW, Low YT, Koh TT, Ithoi I, Mohamed SM (2010) Isolation and characterization of Acanthamoeba spp. from air-conditioners in Kuala Lumpur, Malaysia. Acta Trop 117:23–30. doi:10.1016/j.actatropica.2010.09.004
Chappell CL, Wright JA, Coletta M, Newsome AL (2001) Standardized method of measuring Acanthamoeba antibodies in sera from healthy human subjects. Clin Diagn Lab Immunol 8:724–730. doi:10.1128/CDLI.8.4.724-730.2001
Chomicz L, Piekarczyk J, Starościak B, Fiedor P, Piekarczyk B, Szubińska D, Zawadzki PJ, Walski M (2002) Comparative studies on the occurrence of protozoans, bacteria and fungi in the oral cavity of patients with systemic disorders. Acta Parasitol 47:147–153
Chomicz L, Szaflik JP, Padzik M, Olędzka G, Iwanczyk B, Szaflik J (2012) Anti-amoebic chemotherapeutics and surgical procedures in Acanthamoeba keratitis difficult to diagnose. Proceedings of 47th Congress of the European Society for Surgical Research. Medimond International Proceedings, 55–57
Chomicz L, Padzik M, Graczyk Z, Starościak B, Graczyk T, Naprawska A, Olędzka G, Szostakowska B (2010) Acanthamoeba castellanii: in vitro effects of selected biological, physical and chemical factors. Exp Parasitol 126:103–105. doi:10.1016/j.exppara.2010.01.025
Clarke DW, Niederkorn JY (2006) The pathophysiology of Acanthamoeba keratitis. Trends Parasitol 22:175–180
Corsaro D, Venditti D (2010) Phylogenetic evidence for a new genotype of Acanthamoeba (Amoebozoa, Acanthamoebida). Parasitol Res 107:233–238. doi:10.1007/s00436-010-1870-6
Costa AO, Castro EA, Ferreira GA, Furst C, Crozeta MA, Thomaz-Soccol V (2010) Characterization of Acanthamoeba isolates from dust of a public hospital in Curitiba, Paraná, Brazil. J Eukaryot Microbiol 57:70–75. doi:10.1111/j.1550-7408.2009.00453.x
Edagawa A, Kimura A, Kawabuchi-Kurata T, Kusuhara Y, Karanis P (2009) Isolation and genotyping of potentially pathogenic Acanthamoeba and Naegleria species from tap-water sources in Osaka, Japan. Parasitol Res 105:1109–1117. doi:10.1007/s00436-009-1528-4
Gast RJ, Ledee DR, Fuerst PA, Byers TJ (1996) Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J Eukaryot Microbiol 43:498–504
Gómez-Couso H, Paniagua-Crespo E, Ares-Mazás E (2007) Acanthamoeba as a temporal vehicle of Cryptosporidium. Parasitol Res 100:1151–1154
Gray TB, Cursons RT, Sherwan JF, Rose PR (1995) Acanthamoeba, bacterial, and fungal contamination of contact lens storage cases. Br J Ophthalmol 79:601–605
Hewett MK, Robinson BS, Monis PT, Saint CP (2003) Identification of a new Acanthamoeba 18S rRNA gene sequence type, corresponding to the species Acanthamoeba jacobsi Sawyer, Nerad and Visvesvara, 1992 (Lobosea: Acanthamoebidae). Acta Protozool 42:325–329
Howe DK, Vodkin MH, Novak RJ, Visvesvara G, McLaughlin GL (1997) Identification of two genetic markers that distinguish pathogenic and nonpathogenic strains of Acanthamoeba spp. Parasitol Res 83:345–348
Ibrahim YW, Boase DL, Cree IA (2007) Factors affecting the epidemiology of Acanthamoeba keratitis. Ophthalmic Epidemiol 14:53–60. doi:10.1080/09286580600920281
Kao PM, Hsu BM, Chen NH, Huang KH, Huang SW, King KL, Chiu YC (2012) Isolation and identification of Acanthamoeba species from thermal spring environments in southern Taiwan. Exp Parasitol 130:354–358. doi:10.1016/j.exppara.2012.02.008
Kasprzak W, Mazur T (1972) Free-living amoebae isolated from waters frequented by people in the vicinity of Poznań, Poland. Expeimental studies in mice on the pathogenicity of the isolates. Z Tropenmed Parasitol 23:391–398
Kawaguchi K, Matsuo J, Osaki T, Kamiya S, Yamaguchi H (2009) Prevalence of Helicobacter and Acanthamoeba in natural environment. Lett Appl Microbiol 48:465–471. doi:10.1111/j.1472-765X.2008.02550.x
Khan NA (2006) Acanthamoeba: biology and increasing importance in human health. FEMS Microbiol Rev 30:564–595. doi:10.1111/j.1574-6976.2006.00023.x
Khan NA (2009) Acanthamoeba: biology and pathogenesis. Caister Academic Press, Norfolk, UK
Khan NA, Jarroll EL, Paget TA (2002) Molecular and physiological differentiation between pathogenic and nonpathogenic Acanthamoeba. Curr Microbiol 45:197–202
Khan NA, Jarroll EL, Panjwani J, Cao Z, Paget TA (2000) Proteases as markers for differentiation of pathogenic and nonpathogenic species of Acanthamoeba. J Clin Microbiol 38:2858–2861
Khan NA, Paget TA (2002) Molecular tools for speciation and epidemiological studies of Acanthamoeba. Curr Microbiol 44:444–449
Larkin DF, Kilvington S, Easty DL (1990) Contamination of contact lens storage cases by Acanthamoeba and bacteria. Br J Ophthalmol 74:133–135
Ledee DR, Iovieno A, Miller D, Mandal N, Diaz M, Fell J, Fini ME, Alfonso EC (2009) Molecular identification of T4 and T5 genotypes in isolates from Acanthamoeba keratitis patients. J Clin Microbiol 47:1458–1462. doi:10.1128/JCM.02365-08
Leiva B, Clasdotter E, Linder E, Winiecka-Krusnell J (2008) Free-living Acanthamoeba and Naegleria spp. amebae in water sources of León, Nicaragua. Rev Biol Trop 56:439–446
Lorenzo-Morales J, Lindo JF, Martinez E, Calder D, Figueruelo E, Valladares B, Ortega-Rivas A (2005a) Pathogenic Acanthamoeba strains from water sources in Jamaica, West Indies. Ann Trop Med Parasit 99:751–758
Lorenzo-Morales J, Monteverde-Miranda CA, Jiménez C, Tejedor ML, ValladaresB O-RA (2005b) Evaluation of Acanthamoeba isolates from environmental sources in Tenerife, Canary Islands, Spain. Ann Agric Environ Med 12:233–236
Maghsood AH, Sissons J, Rezaian M, Nolder D, Warhurst D, Khan NA (2005) Acanthamoeba genotype T4 from the UK and Iran and isolation of the T2 genotype from clinical isolates. J Med Microbiol 54:755–759
Magliano ACM, da Silva FM, Teixeira MMG, Alfieri SC (2009) Genotyping, physiological features and proteolytic activities of a potentially pathogenic Acanthamoeba sp. isolated from tap water in Brazil. Exp Parasitol 123:231–235. doi:10.1016/j.exppara.2009.07.006
Mahmoudi MR, Taghipour N, Eftekhar M, Haghighi A, Karanis P (2012) Isolation of Acanthamoeba species in surface waters of Gilan province-north of Iran. Parasitol Res 110:473–477. doi:10.1007/s00436-011-2530-1
Marciano-Cabral F, Cabral G (2003) Acanthamoeba spp. as agents of disease in humans. Clin Microbiol Rev 16:273–307
Niyyati M, Lorenzo-Morales J, Rezaie S, Rahimi F, Mohebali M, Maghsood AH, Motevalli-Haghi A, Martín-Navarro CM, Farnia S, Valladares B, Rezaeian M (2009) Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp Parasitol 121:242–245. doi:10.1016/j.exppara.2008.11.003
Nuprasert W, Putaporntip C, Pariyakanok L, Jongwutiwes S (2010) Identification of a novel T17 genotype of Acanthamoeba from environmental isolates and T10 genotype causing keratitis in Thailand. J Clin Microbiol 48:4636–4640. doi:10.1128/JCM.01090-10
Page FC (1988) A new key to fresh water and soil amoebae. Freshwater Biological Association Scientific Publications, Cumbria England, pp 1–22
Pélandakis M, Pernin P (2002) Use of multiplex PCR and PCR restriction enzyme analysis for detection and exploration of the variability in the free-living amoeba Naegleria in the environment. Appl Environ Microbiol 68:2061–1065. doi:10.1128/AEM.68.4.2061-2065.2002
Pélandakis M, Serre S, Pernin P (2000) Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J Eucaryot Microbiol 47:116–121
Pussard M, Pons R (1977) Morphologies de la paroi kystique et taxonomie du genre Acanthamoeba (Protozoa, Amoebida). Protistologica 13:557–610
Qvarnstrom Y, Visvesvara GS, Sriram R, Da Silva AJ (2006) Multiplex real-time PCR assay for simultaneous detection of Acanthamoeba spp., Balamuthia mandrillaris, and Naegleria fowleri. J Clin Microbiol 44:3589–3595
Rahdar M, Niyyati M, Salehi M, Feghhi M, Makvandi M, Pourmehdi M, Farnia S (2012) Isolation and genotyping of Acanthamoeba strains from environmental sources in Ahvaz City, Khuzestan Province, Southern Iran. Iran J Parasitol 7:22–26
Risler A, Coupat-Goutaland B, Pélandakis M (2013) Genotyping and phylogenetic analysis of Acanthamoeba isolates associated with keratitis. Parasitol Res 112:3807–3816
Rivière D, Szczebara FM, Berjeaud JM, Frère J, Héchard Y (2006) Development of a real-time PCR assay for quantification of Acanthamoeba trophozoites and cysts. J Microbiol Methods 64:78–83
Scheid PL, Schwarzenberger R (2011) Free-living amoebae as vectors of cryptosporidia. Parasitol Res 109:499–504. doi:10.1007/s00436-011-2287-6
Schroeder JM, Booton GC, Hay J, Niszl IA, Seal DV, Markus MB, Fuerst PA, Byers TJ (2001) Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of Acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol 39:1903–1911
Schuster FL, Visvesvara GS (2004) Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int J Parasitol 34:1001–1027
Stockman LJ, Wright CJ, Visvesvara GS, Fields BS, Beach MJ (2011) Prevalence of Acanthamoeba spp. and other free-living amoebae in household water, Ohio, USA—1990–1992. Parasitol Res 108:621–627. doi:10.1007/s00436-010-2120-7
Stothard DR, Schroeder-Diedrich JM, Awwad MH, Gast RJ, Ledee DR, Rodriguez-Zaragoza S, Dean CL, Fuerst PA, Byers TJ (1998) The evolutionary history of the genus Acanthamoeba and the identification of eight new 18s rRNA gene sequence types. J Eukaryot Microbiol 45:45–54
Stratford MP, Griffiths AJ (1978) Variations in the properties and morphology of cysts of Acanthamoeba castellanii. J Gen Microbiol 108:33–37
Szaflik JP, Padzik M, Chomicz L, Olędzka G, Izdebska J, Szaflik J (2012) Usefulness of in vitro diagnostics in difficult incidences of Acanthamoeba keratitis requiring pharmacotherapy and surgical management. Okulistyka 3:28–32
Van Hamme C, Dumont M, Delos M, Lachapelle JM (2001) Cutaneous acanthamoebiasis in a lung transplant patient. Ann Dermatol Venereol 128:1237–1240
Visvesvara GS, Schuster FL (2008) Opportunistic free-living amoebae, part I. Clin Microbiol Newsl 30:151–158
Vodkin MH, Howe DK, Visvesvara GS, McLaughlin GL (1992) Identification of Acanthamoeba at the generic and specific levels using the polymerase chain reaction. J Protozool 39:378–385
Walochnik J, Duchêne M, Seifert K, Obwaller A, Hottkowitz T, Wiedermann G, Eibl H, Aspöck H (2002) Cytotoxic activities of alkylphosphocholines against clinical isolates of Acanthamoeba spp. Antimicrob Agents Ch 46:695–701
Walochnik J, Obwaller A, Aspöck H (2000) Correlations between morphological, molecular biological, and physiological characteristics in clinical and nonclinical isolates of Acanthamoeba spp. Appl Environ Microbiol 66:4408–4413
Wesołowska M, Cisowska A, Myjak P, Marek J, Jurowskaka-Liput J, Jakubaszko J (2006) Acanthamoeba keratitis in contact lens wearers in Poland. Adv Clin Exp Med 15:553–555
Acknowledgments
The authors would like to thank Dr. Alexandre da Silva from the Centers for Disease Control and Prevention, Atlanta, GA, USA, for the Naegleria and Balamuthia strains.
Ethical standards
The manuscript does not contain clinical studies or patient data.
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Lass, A., Szostakowska, B., Idzińska, A. et al. The first genotype determination of Acanthamoeba potential threat to human health, isolated from natural water reservoirs in Poland. Parasitol Res 113, 2693–2699 (2014). https://doi.org/10.1007/s00436-014-3925-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00436-014-3925-6