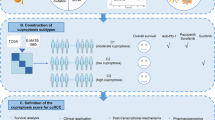
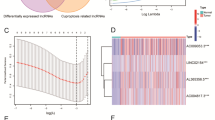
Abstract
Background
Cuproptosis was defined as a novel nonapoptotic cell death pathway and its potential function in clear cell renal cell carcinoma (ccRCC) remains unclear.
Methods
We obtained gene expression profiles, somatic mutation and corresponding clinical information of 881 ccRCC samples from 3 cohorts including the cancer genome atlas cohort, GSE29609 cohort and CheckMate 025 cohort. As described in the latest published article, we enrolled 16 genes as cuproptosis-related genes (CRGs). We explored the expression level, variants and copy number variation of the CRGs. Univariate and multi-variate regression were utilized to assess the prognostic significance of the CRGs. Non-negative matrix factorization was used to identify potential subgroup and gene set variation analysis was used to explore the potential biological functions. CIBERSORT, ESTIMATE algorithm and single sample gene set enrichment analysis were used to evaluate the tumor microenvironment. In vitro experiments including CCK-8, transwell and wound healing assays were utilized to explore the potential biological function of DLAT in ccRCC.
Results
We found that except for CDKN2A, the CRGs were positively associated with patients’ OS. Cuproptosis cluster, cuproptosis gene cluster and cuproptosis score were established, respectively, and higher cuproptosis score was significantly associated with a worse OS in ccRCC (p < 0.001). The area under the receiver operating characteristic curve of the cuproptosis-related nomogram at 1 year, 3 years, 5 years was 0.858, 0.821 and 0.78, respectively. In addition, we found that the cuproptosis score was positively associated with PDCD1, CTLA4 expression level, thus the cuproptosis score may also reflect the dysfunction of tumor infiltrating immune cells. In vitro experiments indicated that overexpression of DLAT could inhibited the migration and proliferation ability of ccRCC cells.
Conclusion
Our findings identify a novel cuproptosis-related signature and the cuproptosis characteristics may influence the anti-tumor immunity though complex regulating networks, and thus cuproptosis may play a role in developing novel therapeutic target of ccRCC.








Similar content being viewed by others
Data availability
Gene expression and corresponding clinical information of 881 ccRCC samples from 3 cohorts as follow: 531 ccRCC samples from TCGA cohort (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga), 39 ccRCC samples from GSE29609 cohort (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), 311 ccRCC samples from patients treated with Everolism or Nivolumab were also obtained from the the online supplemental data (online supplemental table S4) appended to the published paper (https://www.nature.com/articles/s41591-020-0839-y#Sec27).
Abbreviations
- AUC:
-
Area under curve
- ccRCC:
-
Clear cell renal cell carcinoma
- CRGs:
-
Cuproptosis-related genes
- CNV:
-
Copy number variations
- DEG:
-
Differentially expressed genes
- GSVA:
-
Gene set variation analysis
- OS:
-
Overall survival
- PFS:
-
Progression-free survival
- RCC:
-
Renal cell carcinoma
- ROC:
-
Receiver operating characteristic
- ssGSEA:
-
Single sample gene set enrichment analysis
- TCGA:
-
The cancer genome atlas
- TME:
-
Tumor microenvironment
References
Alvarez SW, Sviderskiy VO, Terzi EM, Papagiannakopoulos T, Moreira AL, Adams S, Sabatini DM, Birsoy K, Possemato R (2017) NFS1 undergoes positive selection in lung tumours and protects cells from ferroptosis. Nature 551(7682):639–643
Anderson K, Ryan N, Volpedo G, Varikuti S, Satoskar AR, Oghumu S (2019) Immune suppression mediated by STAT4 deficiency promotes lymphatic metastasis in HNSCC. Front Immunol 10:3095
Braun DA, Hou Y, Bakouny Z, Ficial M, Sant Angelo M, Forman J, Ross-Macdonald P, Berger AC, Jegede OA, Elagina L et al (2020) Interplay of somatic alterations and immune infiltration modulates response to PD-1 blockade in advanced clear cell renal cell carcinoma. Nat Med 26(6):909–918
Dai E, Han L, Liu J, Xie Y, Kroemer G, Klionsky DJ, Zeh HJ, Kang R, Wang J, Tang D (2020) Autophagy-dependent ferroptosis drives tumor-associated macrophage polarization via release and uptake of oncogenic KRAS protein. Autophagy 16(11):2069–2083
Fogel P, Young SS, Hawkins DM, Ledirac N (2007) Inferential, robust non-negative matrix factorization analysis of microarray data. Bioinformatics (Oxf, Engl) 23(1):44–49
Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E et al (2014) Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16(12):1180–1191
Friedmann Angeli JP, Krysko DV, Conrad M (2019) Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer 19(7):405–414
Fu J, Li K, Zhang W, Wan C, Zhang J, Jiang P, Liu XS (2020) Large-scale public data reuse to model immunotherapy response and resistance. Genome Med 12(1):21
Gene Ontology Consortium (2015) Going forward. Nucleic Acids Res 43:1049–1056
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics (Oxf, Engl) 32(18):2847–2849
Guo J, Duan L, He X, Li S, Wu Y, Xiang G, Bao F, Yang L, Shi H, Gao M et al (2021) A combined model of human iPSC-derived liver organoids and hepatocytes reveals ferroptosis in DGUOK Mutant mtDNA depletion syndrome. Adv Sci (Weinh) 8(10):2004680
Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf 14:7
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V (2017) Renal cell carcinoma. Nat Rev Dis Primers 3:17009
Iasonos A, Schrag D, Raj GV, Panageas KS (2008) How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 26(8):1364–1370
Janic A, Valente LJ, Wakefield MJ, Di Stefano L, Milla L, Wilcox S, Yang H, Tai L, Vandenberg CJ, Kueh AJ et al (2018) DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat Med 24(7):947–953
Johnson AM, Kleczko EK, Nemenoff RA (2020) Eicosanoids in cancer: new roles in immunoregulation. Front Pharmacol 11:595498
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30
Kumar P, Bhattacharya P, Prabhakar BS (2018) A comprehensive review on the role of co-signaling receptors and Treg homeostasis in autoimmunity and tumor immunity. J Autoimmun 95:77–99
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxf, Engl) 28(6):882–883
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1(6):417–425
Liu Y, Duan C, Dai R, Zeng Y (2021) Ferroptosis-mediated crosstalk in the tumor microenvironment implicated in cancer progression and therapy. Front Cell Dev Biol 9:739392
Ljungberg B, Cowan NC, Hanbury DC, Hora M, Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF, Sinescu IC (2010) EAU guidelines on renal cell carcinoma: the 2010 update. Eur Urol 58(3):398–406
Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G (2011) GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 12(4):41
Motzer RJ, Molina AM (2009) Targeting renal cell carcinoma. J Clin Oncol 27(20):3274–3276
Motzer RJ, Tannir NM, McDermott DF, Arén Frontera O, Melichar B, Choueiri TK, Plimack ER, Barthélémy P, Porta C, George S et al (2018) Nivolumab plus Ipilimumab versus sunitinib in advanced renal-cell carcinoma. N Engl J Med 378(14):1277–1290
Motzer RJ, Rini BI, McDermott DF, Arén Frontera O, Hammers HJ, Carducci MA, Salman P, Escudier B, Beuselinck B, Amin A et al (2019) Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol 20(10):1370–1385
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12(5):453–457
Powles T, Plimack ER, Soulières D, Waddell T, Stus V, Gafanov R, Nosov D, Pouliot F, Melichar B, Vynnychenko I et al (2020) Pembrolizumab plus axitinib versus sunitinib monotherapy as first-line treatment of advanced renal cell carcinoma (KEYNOTE-426): extended follow-up from a randomised, open-label, phase 3 trial. Lancet Oncol 21(12):1563–1573
Ran X, Xiao J, Zhang Y, Teng H, Cheng F, Chen H, Zhang K, Sun Z (2020) Low intratumor heterogeneity correlates with increased response to PD-1 blockade in renal cell carcinoma. Ther Adv Med Oncol 12:1758835920977117
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47
Sheftel AD, Stehling O, Pierik AJ, Elsässer HP, Mühlenhoff U, Webert H, Hobler A, Hannemann F, Bernhardt R, Lill R (2010) Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc Natl Acad Sci U S A 107(26):11775–11780
Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, Brown LM, Valenzuela CA, Wolpaw AJ, Stockwell BR (2016) Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol 12(7):497–503
Strasser A, Vaux DL (2020) Cell death in the origin and treatment of cancer. Mol Cell 78(6):1045–1054
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, Rossen J, Joesch-Cohen L, Humeidi R, Spangler RD et al (2022) Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375(6586):1254–1261
van HoogstadEvert JS, Bekkers R, Ottevanger N, Jansen JH, Massuger L, Dolstra H (2020) Harnessing natural killer cells for the treatment of ovarian cancer. Gynecol Oncol 157(3):810–816
White E, Mehnert JM, Chan CS (2015) Autophagy, metabolism, and cancer. Clin Cancer Res 21(22):5037–5046
Wong RS (2011) Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res 30(1):87
Woo SM, Seo SU, Min KJ, Im SS, Nam JO, Chang JS, Kim S, Park JW, Kwon TK (2018) Corosolic acid induces non-apoptotic cell death through generation of lipid reactive oxygen species production in human renal carcinoma caki cells. Int J Mol Sci 19:5
Ye J, Jiang X, Dong Z, Hu S, Xiao M (2019) Low-concentration PTX And RSL3 inhibits tumor cell growth synergistically by inducing ferroptosis in mutant p53 hypopharyngeal squamous carcinoma. Cancer Manag Res 11:9783–9792
Acknowledgements
We thank the TCGA, GEO database for providing NGS data and clinical information of ccRCC.
Funding
This work is supported by Grants from the National Key Research and Development Program of China (No. 2019YFC1316005), National Natural Science Foundation of China (Nos. 81772706, 81802525 and 81902568), Shanghai Science and Technology Committee (Nos. 20ZR1413100, 18511108000), and Shanghai Sailing Program (No. 19YF1409700).
Author information
Authors and Affiliations
Contributions
The work reported in the paper has been performed by the authors, unless clearly specified in the text. Conceptualization: XT, SZ, WX, GW, and CC. Data curation and formal analysis: XT, WL, SZ, XW, WX, AA, GW, and JZ. Funding acquisition: HZ and DY. Investigation and methodology: XT, XW, WL, WX, AA, SY and XC. Resources and software: WX, YQ, HZ and DY. Supervision: GW, YQ, HZ and DY. Validation and visualization: WX, AA and WL. Original draft: XT, SZ, CC, XW, and GW. Editing: WX, YQ, HZ and DY.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare no competing interests.
Ethics statement
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tian, X., Zhu, S., Liu, W. et al. Construction of cuproptosis signature based on bioinformatics and experimental validation in clear cell renal cell carcinoma. J Cancer Res Clin Oncol 149, 17451–17466 (2023). https://doi.org/10.1007/s00432-023-05259-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-023-05259-z