Abstract
Purpose
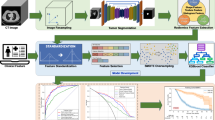
To evaluate a new radiomics strategy that incorporates intratumoral and peritumoral features extracted from lung CT images with ensemble learning for pretreatment prediction of lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD).
Methods
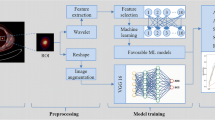
A total of 105 patients (47 LUSC and 58 LUAD) with pretherapy CT scans were involved in this retrospective study, and were divided into training (n = 73) and testing (n = 32) cohorts. Seven categories of radiomics features involving 3078 metrics in total were extracted from the intra- and peritumoral regions of each patient’s CT data. Student’s t tests in combination with three feature selection methods were adopted for optimal features selection. An ensemble classifier was developed using five common machine learning classifiers with these optimal features. The performance was assessed using both training and testing cohorts, and further compared with that of Visual Geometry Group-16 (VGG-16) deep network for this predictive task.
Results
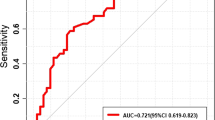
The classification models developed using optimal feature subsets determined from intratumoral region and peritumoral region with the ensemble classifier achieved mean area under the curve (AUC) of 0.87, 0.83 in the training cohort and 0.66, 0.60 in the testing cohort, respectively. The model developed by using the optimal feature subset selected from both intra- and peritumoral regions with the ensemble classifier achieved great performance improvement, with AUC of 0.87 and 0.78 in both cohorts, respectively, which are also superior to that of VGG-16 (AUC of 0.68 in the testing cohort).
Conclusions
The proposed new radiomics strategy that extracts image features from the intra- and peritumoral regions with ensemble learning could greatly improve the diagnostic performance for the histological subtype stratification in patients with NSCLC.









Similar content being viewed by others
Data availability statement
The raw/processed data of this study cannot be publicly shared at present as it forms part of an ongoing study, but it could be available under reasonable request from the corresponding author with the permission of the Institutional Review Board. Results and code package in each step of this study have been arranged in a document named as “Appendix”. The code package has also been uploaded to Gitee for publicly sharing and further perfection (https://gitee.com/yang-tianran-01/radiomics_-ensemble_learning/commit/d51e6859ef48c92cc0c794639f08286ac89569f8).
Abbreviations
- AUC:
-
Area under the curve
- CM:
-
Co-occurrence matrices
- CNN:
-
Convolutional neural network
- CT:
-
Computed tomography
- FN:
-
False negative
- FP:
-
False positive
- GLCM:
-
Gray-level co-occurrence matrix
- GLDM:
-
Gray-level dependence matrix
- GLRLM:
-
Gray-level run length matrix
- GLSZM:
-
Gray-level size zone matrix
- LASSO:
-
Least absolute shrinkage and selection operator
- LBP:
-
Local binary pattern
- LUAD:
-
Lung adenocarcinoma
- LUSC:
-
Lung squamous cell carcinoma
- MID:
-
Mutual information difference
- mpMRI:
-
Multiparametric magnetic resonance imaging
- mRMR:
-
Minimum redundancy maximum relevance
- NGTDM:
-
Neighboring gray-tone difference matrix
- NSCLC:
-
Nonsmall-cell lung cancer
- PET-CT:
-
Positron emission tomography computed tomography
- QDA:
-
Quadratic discriminant analysis
- RBF:
-
Radial basis function
- RF:
-
Random forest
- RLM:
-
Run length matrix
- ROC:
-
Receiver-operating characteristic curve
- SVM:
-
Support vector machine
- SVM-RFE:
-
Support vector machine-based recursive feature elimination
- TN:
-
True negative
- TP:
-
True positive
- VGG:
-
Visual geometry group network
- XGBoost:
-
Extreme gradient boosting
References
Amadasun M, King R (1989) Texural features corresponding to texural properties. IEEE Trans Syst Man Cybern 19(5):1264–1274
Bashir U et al (2019) Non-invasive classifcation of non-small cell lung cancer: a comparison between random forest models utilising radiomic and semantic features. Br J Radiol 92(20190159):1–8
Beig N et al (2019) Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology 290(3):783–792
Bray F et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. https://doi.org/10.3322/caac.21492
Chaunzwa TL et al (2018) Using deep-learning radiomics to predict lung cancer histology. J Clin Oncol 36(15_Suppl):8545–8545
Chen T, Guestrin C (2016) XGBoost: a scalable tree boosting system. In: 2016. Association for Computing Machinery
Colen RR et al (2021) Radiomics analysis for predicting pembrolizumab response in patients with advanced rare cancers. J Immunother Cancer 9(4):1752
de Jong EEC et al (2018) Applicability of a prognostic CT-based radiomic signature model trained on stage I-III non-small cell lung cancer in stage IV non-small cell lung cancer. Lung Cancer 124:6–11
Ebrahimi M et al (2016) Diagnostic concordance of non–small cell lung carcinoma subtypes between biopsy and cytology specimens obtained during the same procedure. Cancer Cytopathol 124(10):737–743
Fehr D et al (2015) Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci 112(46):E6265–E6273
Galloway MM (1975) Texture analysis using gray level run lengths. Comput Graph Image Process 4:172–179
Gupta V, Mittal M (2019a) R-peak detection in ECG signal using yule-walker and principal component analysis. IETE J Res 67:921–934
Gupta V, Mittal M (2019b) A comparison of ECG signal pre-processing using FrFT, FrWT and IPCA for improved analysis. IRBM 40:145–156
Gupta V, Mittal M (2019c) QRS complex detection using STFT, chaos analysis, and PCA in standard and real-time ECG databases. J Inst Eng (india) 100:489–497
Haralick RM, Shanmugam K, Dinstein IH (1973) Textural features for image classification. IEEE Trans Syst Man Cybern 3(6):610–621
Hastie T, Tibshirani R, Friedman J (2009) The elements of statistical learning: data mining, inference, and prediction, 2nd edn. Springer Science & Business Media, Berlin, p 757
Herbst RS, Heymach JV, Lippman SM (2008) Lung cancer. N Engl J Med 359(13):1367–1380
Hoffman PC, Mauer AM, Vokes EE (2000) Lung cancer. Lancet 355(9202):479–485
Khalilia M, Chakraborty S, Popescu M (2011) Predicting disease risks from highly imbalanced data using random forest. BMC Med Inform Decis Mak 11(1):51
Kirienko M et al (2018) Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging 45(2):207–217
Kora P, Krishna KSR (2014) Myocardial infarction detection using magnitude squared coherence and support vector machine. Med Imaging. https://doi.org/10.1109/MedCom.2014.7006037
Koyasu S et al (2020) Usefulness of gradient tree boosting for predicting histological subtype and EGFR mutation status of non-small cell lung cancer on 18F FDG-PET/CT. Ann Nucl Med 34(1):49–57
Lam H-K et al (2012) Computational intelligence and its applications: evolutionary computation, fuzzy logic, neural network and support vector machine techniques. World Scientific, London, p 318
Li S et al (2019) Predicting lung nodule malignancies by combining deep convolutional neural network and handcrafted features. Phys Med Biol 64(17):175012
Liang C et al (2018) A computer-aided diagnosis scheme of breast lesion classification using GLGLM and shape features: Combined-view and multi-classifiers. Phys Med 55:61–72
Linear & Quadratic Discriminant Analysis · UC Business Analytics R Programming Guide. https://uc-r.github.io/discriminant_analysis
Ma Y et al (2018a) Intra-tumoural heterogeneity characterization through texture and colour analysis for differentiation of non-small cell lung carcinoma subtypes. Phys Med Biol 63(16):1658
Ma Y et al (2018b) Intra-tumoural heterogeneity characterization through texture and colour analysis for differentiation of non-small cell lung carcinoma subtypes. Phys Med Biol 63(16):1658
Mahon RN, Hugo GD, Weiss E (2019) Repeatability of texture features derived from magnetic resonance and computed tomography imaging and use in predictive models for non-small cell lung cancer outcome. Phys Med Biol 64:145007
Peng H, Long F, Ding C (2005) Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 27(8):1226–1238
Probable T (1992) Error of a mean. In: Kotz S, Johnson NL (eds) Breakthroughs in statistics: methodology and distribution. Springer, New York, pp 33–57
Ren C et al (2020) Machine learning based on clinico-biological features integrated 18F-FDG PET/CT radiomics for distinguishing squamous cell carcinoma from adenocarcinoma of lung. Eur J Nucl Med Mol Imaging. https://doi.org/10.1007/s00259-020-05065-6
Sauerbrei W, Royston P, Binder H (2007) Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat Med 26(30):5512–5528
Seera M, Lim CP (2014) A hybrid intelligent system for medical data classification. Expert Syst Appl 41(5):2239–2249
Shen C et al (2017) 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl Oncol 10(6):886–894
Sollini M et al (2017) PET Radiomics in NSCLC: state of the art and a proposal for harmonization of methodology. Sci Rep 7(1):358
Starkov P et al (2018) The use of texture-based radiomics CT analysis to predict outcomes in early-stage non-small cell lung cancer treated with stereotactic ablative radiotherapy. Br J Radiol 91(20180228):1–7
Stenzinger A et al (2021) Artificial intelligence and pathology: from principles to practice and future applications in histomorphology and molecular profiling. Semin Cancer Biol. https://doi.org/10.1016/j.semcancer.2021.02.011
Su R et al (2019) Identification of expression signatures for non-small-cell lung carcinoma subtype classification. Bioinformatics. https://doi.org/10.1093/bioinformatics/btz557
Sun C, Wee WG (1983) Neighboring gray level dependence matrix for texture classification. Compute vis Graph Image Process 23:341–352
Sun W et al (2018) Effect of machine learning methods on predicting NSCLC overall survival time based on Radiomics analysis. Radiat Oncol 13(1):197
Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin 71:209–249
Tang X et al (2020) Elaboration of a multimodal MRI-based radiomics signature for the preoperative prediction of the histological subtype in patients with non-small-cell lung cancer. BioMed Eng Online. https://doi.org/10.1186/s12938-019-0744-0
Tharwat A (2016) Linear vs quadratic discriminant analysis classifier: a tutorial. Int J Appl Pattern Recogn 3(2):145
Thibault G, Angulo J, Meyer F (2014) Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng 61(3):630–637
Tibshirani R (1996) Regression Shrinkage and Selection Via the Lasso. J Roy Stat Soc Ser B (methodol) 58(1):267–288
van Griethuysen JJM et al (2017) Computational radiomics system to decode the radiographic phenotype. Can Res 77(21):e104–e107
Wu W et al (2016) Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol. https://doi.org/10.3389/fonc.2016.00071
Wu S et al (2017) A radiomics nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. Clin Cancer Res 23(22):6904–6911
Wu S et al (2018) Development and validation of an MRI-based radiomics signature for the preoperative prediction of lymph node metastasis in bladder cancer. EBioMedicine 34:76–84
Xu X et al (2017a) Preoperative prediction of muscular invasiveness of bladder cancer with radiomic features on conventional MRI and its high-order derivative maps. Abdom Radiol (NY) 42(7):1896–1905
Xu X et al (2017b) Three-dimensional texture features from intensity and high-order derivative maps for the discrimination between bladder tumors and wall tissues via MRI. Int J Comput Assist Radiol Surg 12(4):645–656
Xu X et al (2019) A predictive nomogram for individualized recurrence stratification of bladder cancer using multiparametric MRI and clinical risk factors. J Magn Resonance Imaging 50:1893–1904
Zhu X et al (2018) Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur Radiol 28(7):1–7
Zwanenburg A et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295(2):328–338
Funding
This work was funded by the National Natural Science Foundation of China (No. 81901698) and Young Eagle plan of High Ambition Project (No. 2020CYJHXXP).
Author information
Authors and Affiliations
Contributions
XX, XT, and HH contributed to the study concept, design, and data interpretation. XT contributed to the CT and clinical data collection. XT and HY contributed to the intratumoral region annotation. HH, XX and PD performed the peritumoral region extraction and radiomics feature calculation; XX, HH and XT contributed to the model construction and data analysis. XX, XT, and HH contributed to the manuscript drafting, editing and revision. All authors approve the final version of the manuscript for submission.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the institutional ethics review board of Xijing Hospital, and informed content was waived.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tang, X., Huang, H., Du, P. et al. Intratumoral and peritumoral CT-based radiomics strategy reveals distinct subtypes of non-small-cell lung cancer. J Cancer Res Clin Oncol 148, 2247–2260 (2022). https://doi.org/10.1007/s00432-022-04015-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-022-04015-z