Abstract
Objective
Clear cell renal cell carcinoma (ccRCC) is the most common type of kidney tumor characterized by the highest mortality rate of the genitourinary cancers, and, therefore, new diagnostic and/or prognostic biomarkers are urgently needed.
Methods
Based on genome-wide DNA methylation profiling in 11 pairs of ccRCC and non-cancerous renal tissues (NRT), the methylation at regulatory regions of ZNF677, FBN2, PCDH8, TFAP2B, TAC1, and FLRT2 was analyzed in 168 renal tissues and 307 urine samples using qualitative and quantitative methylation-specific PCR (MSP).
Results
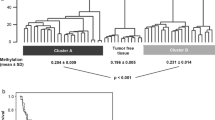
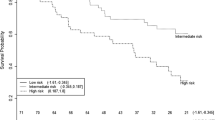
Significantly higher methylation frequencies for all genes were found in ccRCC tissues compared to NRT (33–60% vs. 0–11%). The best diagnostic performance demonstrated a panel of ZNF677, FBN2, PCDH8, TFAP2B & TAC1 with 82% sensitivity and 96% specificity. Hypermethylation of ZNF677 and PCDH8 in the tissue samples was significantly related to numerous adverse clinicopathologic parameters. For the urine-based ccRCC detection, the highest diagnostic power (AUC = 0.78) was observed for a panel of ZNF677 & PCDH8 (with or without FBN2 or FLRT2) with 69–78% sensitivity and 69–80% specificity, albeit with lower values in the validation cohort. Besides, methylation of PCDH8 was significantly related to higher tumor stage and fat invasion in the study and validation cohorts. Moreover, PCDH8 was strongly predictive for OS (HR, 5.7; 95% CI 1.16–28.12), and its prognostic power considerably increased in combination with ZNF677 (HR, 12.5; 95% CI 1.47–105.58).
Conclusion
In summary, our study revealed novel, potentially promising DNA methylation biomarkers of ccRCC with the possibility to be applied for non-invasive urine-based ccRCC detection and follow-up.






Similar content being viewed by others
Availability of data and materials
All data supporting the results reported in the article is available from the corresponding author upon a reasonable request.
Abbreviations
- AUC:
-
Area under the curve
- ASC:
-
Asymptomatic control
- ccRCC:
-
Clear cell renal cell carcinoma
- CI:
-
Confidence interval
- MC:
-
In vitro methylated control
- MSP:
-
Methylation-specific polymerase chain reaction
- qMSP:
-
Quantitative methylation-specific polymerase chain reaction
- NRT:
-
Non-cancerous renal tissue
- NTC:
-
No-template control
- HR:
-
Hazard ratio
- UC:
-
Unmethylated control
- M:
-
Methylated
- U:
-
Unmethylated
- ROC:
-
Receiver operating characteristic
- AUC:
-
Area under the curve
- DSe:
-
Diagnostic sensitivity
- DSp:
-
Diagnostic specificity
- WHO/ISUP:
-
World health organization/International Society of Urological Pathology
References
Bae H, Kim B, Lee H, Lee S, Kang HS, Kim SJ (2017) Epigenetically regulated Fibronectin leucine rich transmembrane protein 2 (FLRT2) shows tumor suppressor activity in breast cancer cells. Sci Rep 7(1):272
Bakavicius A, Daniunaite K, Zukauskaite K, Barisiene M, Jarmalaite S, Jankevicius F (2019) Urinary DNA methylation biomarkers for prediction of prostate cancer upgrading and upstaging. Clin Epigenet 11(1):115
Battagli C, Uzzo RG, Dulaimi E, Ibanez de Caceres I, Krassenstein R, Al-Saleem T et al (2003) Promoter hypermethylation of tumor suppressor genes in urine from kidney cancer patients. Cancer Res 63:8695–8699
Bedke J, Buse S, Pritsch M, Macher-Goeppinger S, Schirmacher P, Haferkamp A et al (2009) Perinephric and renal sinus fat infiltration in pT3a renal cell carcinoma: possible prognostic differences. BJU Int 103(10):1349–1354
Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L et al (2015) STARD Group. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 351:h5527
Capitanio U, Bensalah K, Bex A et al (2019) Epidemiology of renal cell carcinoma. Eur Urol 75(1):74–84
Chow WH, Dong LM, Devesa SS (2010) Epidemiology and risk factors for kidney cancer. Nat Rev Urol 7:245–257
Costa VL, Henrique R, Danielsen SA, Knaes M, Patrício P, Morais A et al (2011) TCF21 and PCDH17 methylation: an innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics 6:1120–1130
Daniunaite K, Jarmalaite S, Kalinauskaite N, Petroska D, Laurinavicius A, Lazutka JR et al (2014) Prognostic value of RASSF1 promoter methylation in prostate cancer. J Urol 192(6):1849–1855
Daniunaite K, Serenaite I, Misgirdaite R, Gordevicius J, Unguryte A, Fleury-Cappellesso S et al (2015) Epigenetic regulation of human adipose-derived stem cells differentiation. Mol Cell Biochem 410:111–120
Daniūnaitė K, Jarmalaitė S, Kriukienė E (2019) Epigenomic technologies for deciphering circulating tumor DNA. Curr Opin Biotechnol 55:23–29
Delahunt B, Cheville JC, Martignoni G, Humphrey PA, Magi-Galluzzi C, McKenney J et al (2013) The International Society of Urological Pathology (ISUP) grading system for renal cell carcinoma and other prognostic parameters. Am J Surg Pathol 37:1490
Eckert D, Buhl S, Weber S, Jäger R, Schorle H (2005) The AP-2 family of transcription factors. Genome Biol 6(13):246
Flintoff KA, Arudchelvan Y, Gong SG (2014) FLRT2 interacts with fibronectin in the ATDC5 chondroprogenitor cells. J Cell Physiol 229(10):1538–1547
Fuhrman SA, Lasky LC, Limas C (1982) Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol 6:655–663
Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I et al (2014) Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet 46(3):225–233
Heller G, Altenberger C, Schmid B, Marhold M, Tomasich E, Ziegler B et al (2015) DNA methylation transcriptionally regulates the putative tumor cell growth suppressor ZNF677 in non-small cell lung cancers. Oncotarget 6:394–408
Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH et al (2004) Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res 64(15):5511–5517
Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M et al (2017a) Renal cell carcinoma. Nat Rev Dis Primers 3:17009
Hsieh JJ, Manley BJ, Khan N, Gao JJ, Carlo MI, Cheng EH (2017b) Overcome tumor heterogeneity-imposed therapeutic barriers through convergent genomic biomarker discovery: a braided cancer river model of kidney cancer. Semin Cell Dev Biol 64:98–106
Hu CY, Mohtat D, Yu Y, Ko YA, Shenoy N, Bhattacharya S et al (2014) Kidney cancer is characterized by aberrant methylation of tissue-specific enhancers that are prognostic for overall survival. Clin Cancer Res 20(16):4349–4360
Jayson M, Sanders H (1998) Increased incidence of serendipitously discovered renal cell carcinoma. Urology 51(2):203–205
Kabaria R, Klaassen Z, Terris MK (2016) Renal cell carcinoma: links and risks. Int J Nephrol Renovasc Dis 9:45–52
Kubiliute R, Jarmalaite S (2021) Epigenetic biomarkers of renal cell carcinoma for liquid biopsy tests. Int J Mol Sci 22(16):8846
Larsen LK, Lind GE, Guldberg P, Dahl Ch (2019) DNA-methylation-based detection of urological cancer in urine: overview of biomarkers and considerations on biomarker design, source of DNA, and detection technologies. Int J Mol Sci 20(11):2657
Lasseigne BN, Brooks JD (2018) The role of DNA methylation in renal cell carcinoma. Mol Diagn Ther 22(4):431–442
Lehmann U, Langer F, Feist H, Glöckner S, Hasemeier B, Kreipe H (2002) Quantitative assessment of promoter hypermethylation during breast cancer development. Am J Pathol 160(2):605–612
Li Y, Yang Q, Guan H, Shi B, Ji M, Hou P (2018) ZNF677 suppresses Akt phosphorylation and tumorigenesis in thyroid cancer. Cancer Res 78(18):5216–5228
Li L, Shi L, Zhang J, Fan Y, Li Q (2021) The critical impact of tumor size in predicting cancer special survival for T3aM0M0 renal cell carcinoma: A proposal of an alternative T3aN0M0 stage. Cancer Med 10(2):605–614
Lin YL, Wang YL, Fu XL, Ma JG (2014) Aberrant methylation of PCDH8 is a potential prognostic biomarker for patients with clear cell renal cell carcinoma. Med Sci Monit 20:2380–2385
Lin Y, Ge X, Zhang X, Wu Z, Liu K, Lin F et al (2018) Protocadherin-8 promotes invasion and metastasis via laminin subunit γ2 in gastric cancer. Cancer Sci 109(3):732–740
Lopez-Beltran A, Scarpelli M, Montironi R, Kirkali Z (2006) 2004 WHO classification of the renal tumors of the adults. Eur Urol 49:798–805
Lu T, Li J (2017) Clinical applications of urinary cell-free DNA in cancer: current insights and promising future. Am J Cancer Res 7(11):2318–2332
Maleckaite R, Zalimas A, Bakavicius A, Jankevicius F, Jarmalaite S, Daniunaite K (2019) DNA methylation of metallothionein genes is associated with the clinical features of renal cell carcinoma. Oncol Rep 41(6):3535–3544
McMahon KW, Karunasena E, Ahuja N (2017) The roles of DNA methylation in the stages of cancer. Cancer J 23(5):257–261
Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H et al (2011) Genome-wide methylation analysis identifies epigenetically inactivated candidate tumor suppressor genes in renal cell carcinoma. Oncogene 30(12):1390–1401
Moser M, Pscherer A, Roth C, Becker J, Mücher G, Zerres K et al (1997) Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2beta. Genes Dev 11(15):1938–1948
Neuzillet C, Tijeras-Raballand A, Cohen R, Cros J, Faivre S, Raymond E et al (2015) Targeting the TGFbeta pathway for cancer therapy. Pharmacol Ther 147:22–31
Outeiro-Pinho G, Barros-Silva D, Aznar E, Sousa AI, Vieira-Coimbra M, Oliveira J et al (2020) MicroRNA-30a-5pme: a novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J Exp Clin Cancer Res 39(1):98
Pires-Luís AS, Costa-Pinheiro P, Ferreira MJ, Antunes L, Lobo F, Oliveira J et al (2017) Identification of clear cell renal cell carcinoma and oncocytoma using a three-gene promoter methylation panel. J Transl Med 15:149
Rameshwar P, Gascón P (1996) Induction of negative hematopoietic regulators by neurokinin-A in bone marrow stroma. Blood 88(1):98–106
Ricketts CJ, Hill VK, Linehan WM (2014) Tumor-specific hypermethylation of epigenetic biomarkers, including SFRP1, predicts for poorer survival in patients from the TCGA Kidney Renal Clear Cell Carcinoma (KIRC) project. PLoS ONE 9(1):e85621
Rossi SH, Prezzi D, Kelly-Morland C, Goh V (2018) Imaging for the diagnosis and response assessment of renal tumours. World J Urol 36(12):1927–1942
Sauerbrei W, Taube SE, McShane LM, Cavenagh MM, Altman DG (2018) Reporting recommendations for tumor marker prognostic studies (REMARK): an abridged explanation and elaboration. J Natl Cancer Inst 110(8):803–811
Siegel RL, Miller KD, Jemal A (2015) Cancer statistics, 2015. CA Cancer J Clin 65:5–29
Sobin LH, Gospodarowicz MK, Wittekind C (eds) (2009) TNM classification of malignant tumours, 7th Edition. UICC International Union Against Cancer. 7:310
Sohlberg EM, Metzner TJ, Leppert JT (2019) The harms of overdiagnosis and overtreatment in patients with small renal masses: A mini-review. Eur Urol Focus 5(6):943–945
The Cancer Genome Atlas Research Network (2013) Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499(7456):43–49
Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T et al (2018a) Deterministic evolutionary trajectories influence primary tumor growth: TRACERx renal. Cell 173:595–610
Turajlic S, Swanton Ch, Boshoff Ch (2018b) Kidney cancer: The next decade. J Exp Med 215(10):2477–2479
van Loon K, Yemelyanenko-Lyalenko J, Margadant C, Griffioen AW, Huijbers EJM (2020) Role of fibrillin-2 in the control of TGF-β activation in tumor angiogenesis and connective tissue disorders. Biochim Biophys Acta Rev Cancer 1873(2):188354
van Vlodrop IJ, Baldewijns MM, Smits KM, Schouten LJ, van Neste L, van Criekinge W et al (2010) Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma. Am J Pathol 176(2):575–584
van Vlodrop IJH, Joosten SC, De Meyer T, Smits KM, Van Neste L, Melotte V et al (2017) A four-gene promoter methylation marker panel consisting of GREM1, NEURL, LAD1, and NEFH predicts survival of clear cell renal cell cancer patients. Clin Cancer Res 23(8):2006–2018
Warren AY, Harrison D (2018) WHO/ISUP classification, grading and pathological staging of renal cell carcinoma: standards and controversies. World J Urol 36(12):1913–1926
Williamson SR, Taneja K, Cheng L (2019) Renal cell carcinoma staging: pitfalls, challenges, and updates. Histopathology 74(1):18–30
Yu JS, Koujak S, Nagase S, Li CM, Su T, Wang X et al (2008) PCDH8, the human homolog of PAPC, is a candidate tumor suppressor of breast cancer. Oncogene 27:4657–4665
Acknowledgements
The authors would like to thank Aušra Šumskaitė, Rūta Matuliavičiūtė, and Eugenijus Ganža for their assistance in DNA samples preparation and MSP analysis.
Funding
This work was funded by the 2014–2020 European Union Structural Funds according to the activity "Intelligence. Joint science-business projects" grant No. J05-LVPA-K-04–0029. SJ and KZ were supported by the European Social Fund according to the activity "Development of students' ability to carry out R&D activities" under Measure No.09.03.3-LMT-K-712 "Development of Scientific Competences of Scientists, other Researchers and Students through Practical Research Activities" (grant No. 09.03.3-LMT-K-712–15-0214 to SJ). The study was also partially supported by the Research Council of Lithuania (RCL) grant No. S-MIP-17/54.
Author information
Authors and Affiliations
Contributions
RK: performed the DNA methylation and gene expression analysis in the tissue samples and urine samples of the validation cohort, analyzed the experimental and clinical data and drafted the manuscript; KZ: performed the DNA methylation analysis in urine samples of the study cohort, contributed to gene expression analysis; AZ, AU, RS and AB: collected the clinical data and revised the analysis; RS and AZ: critically revised the manuscript; AU and FJ: coordinated the patient selection, supervised the clinical data analysis and was involved in the conception of the study; SJ: designed the research, supervised the analysis of the clinical and experimental data, and revised the manuscript critically for important intellectual content. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work is appropriately investigated and resolved.
Corresponding author
Ethics declarations
Competing interests
RK and SJ are inventors of the patent application No. PCT/IB2021/052532. The remaining authors declare that they have no conflict of interest.
Ethics approval and consent to participate
The study was approved by the Lithuanian Bioethics Committee (Nr. 158200˗18/12˗1077˗585 for Study cohort and Nr. 158200-18/12-1077-585 for Validation cohort), and written informed consent was obtained from all patients.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kubiliūtė, R., Žukauskaitė, K., Žalimas, A. et al. Clinical significance of novel DNA methylation biomarkers for renal clear cell carcinoma. J Cancer Res Clin Oncol 148, 361–375 (2022). https://doi.org/10.1007/s00432-021-03837-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00432-021-03837-7