Abstract
Although the bys-like family of genes has been conserved from yeast to humans, it is not apparent to what extent the function of Bys-like proteins has been conserved across phylogenetic groups. Human Bystin is thought to function in a novel cell adhesion complex involved in embryo implantation. The product of the yeast bys-like gene, Enp1, is nuclear and has a role in pre-ribosomal RNA (pre-rRNA) splicing and ribosome biogenesis. To gain insight into the function of the Drosophila melanogaster bys-like family member, termed bys, we examined bys mRNA expression and the localization of Bys protein. In embryos, bys mRNA is expressed in a tissue-specific pattern during gastrulation. In the larval wing imaginal disc, bys mRNA is expressed in the ventral and dorsal regions of the wing pouch, regions that give rise to epithelia that adhere to one another after the wing disc everts. The bys mRNA expression patterns could be interpreted as being consistent with a role for Bys in events requiring cell-cell interactions. However, embryonic bys mRNA expression patterns mirror those of genes that are potential targets of the growth regulator Myc and encode nucleolar proteins implicated in cell growth. Additionally, in Schneider line 2 (S2) cells, an epitope-tagged Bys protein is localized to the nucleus, suggesting that Drosophila Bys function may be conserved with that of yeast Enp1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The human proteins Bystin, Trophinin and Tastin were identified in a search for proteins that function in cell adhesion during embryo implantation. Database searches revealed that Bystin was similar to the protein encoded by the previously reported bys gene in Drosophila melanogaster, but proteins similar to Trophinin and Tastin were not found in Drosophila. Based on their amino acid sequences, Bystin and Tastin were predicted to be cytoplasmic while Trophinin was predicted to be an integral membrane protein. Yeast two-hybrid studies showed that Bystin interacts with Trophinin, Tastin and cytokeratins, but that Trophinin and Tastin do not directly associate with one another. Bystin and Tastin were shown to be cytoplasmic and it was demonstrated that Bystin, Tastin and Trophinin were not expressed in most human tissues, but were co-expressed in the trophoblast and endometrial epithelium at the place where embryo implantation had occurred. These studies led to the suggestion that Bystin, Trophinin and Tastin may function in a complex to mediate homophilic cell adhesion during human embryo implantation (reviewed in Aoki and Fukuda 2000 and references therein).
However, work reported with mice suggested that Bystin and Trophinin are not necessary for implantation in mice, but may be important for early development (Suzuki et al. 1999). The expression pattern of mouse Trophinin was not consistent with a role for the protein in implantation. Additionally, in one genetic background, embryos lacking the trophinin gene did implant into the uteri of trophinin± heterozygous females, but showed partial embryonic lethality (Nadano et al. 2002). In the trophinin knock-out study, the authors reported as unpublished data that bystin knock-out mice showed completely penetrant lethality after implantation (Nadano et al. 2002).
In the yeast Saccharomyces cerevisiae, the ENP1 gene is required for vegetative growth and encodes a protein with similarity to the mammalian Bystin proteins and Drosophila Bys. However, the yeast Enp1 protein is localized to the nucleus, enriched in the nucleolus and plays a role in ribosomal RNA (rRNA) processing and ribosome biogenesis (Roos et al. 1997; Chen et al 2003). At the non-permissive temperature, yeast with a temperature-sensitive mutation in ENP1 had reduced levels of 40S ribosomal subunits and defective splicing of the 20S pre-ribosomal RNA (pre-rRNA), a precursor of the 18S rRNA (Chen et al. 2003). In another study, Enp1 was found as part of a 90S pre-ribosomal complex and associated strongly with 20S pre-rRNA. In this study, Enp1 was found in the nucleolus of most cells, but was present in the cytoplasm of some cells. Because Enp1 strongly associated with the 20S pre-rRNA, which is usually exported from the nucleus as part of pre-40S ribosomes, it was suggested that Enp1 may accompany pre-40S ribosomes to the cytoplasm (Grandi et al. 2002).
During studies of yeast ENP1, it was noted that the human bystin cDNA that was initially identified in cell adhesion assays and used for protein-protein interaction studies was incomplete. When a longer cDNA coding for a presumptive full-length Bystin of 437 amino acids was tagged at the N-terminus with GFP and expressed in yeast, Bystin was found in the nucleus and enriched in the nucleolus. Based on these observations and the function of ENP1, it was proposed that the conserved role of Bys-like proteins is in rRNA processing. However, human Bystin did not complement a yeast enp1 null mutant (Chen et al. 2003), indicating that human Bystin does not functionally substitute for ENP1. In light of the different potential functions of human Bystin, mouse Bystin and yeast Enp1, we asked if the Drosophila bys mRNA expression patterns or the sub-cellular location of Bys protein would suggest that Drosophila Bys function might be similar to that of previously described Bys-like proteins.
Materials and methods
Whole-mount in situ hybridization
Antisense and sense digoxygenin-labeled RNA probes were made using a bys cDNA coding for the full-length Bys protein and a DIG RNA labeling kit (Roche). In situ hybridizations to ovaries, embryos and discs were performed as described (Lehmann and Tautz 1994; O’Neill and Bier 1994; Deng et al. 1999). Embryos were mounted in Canada balsam:methylsalicylate (6:1). Ovaries and discs were mounted in 80% glycerol. Specimens were photographed with a Spot digital camera.
Generation of myc-tagged Bys
A sense oligonucleotide primer was designed with two regions complementary to a bys cDNA flanking a sequence coding for the myc epitope C-terminus. The sense primer was 5′-GACAATATCCGAAA aagcttatctccgaggaggacctGGGTAAGCCGAAGAAGGC-3′. The region coding for the C-terminus of the myc epitope is in lower case and a HindIII site within the myc epitope is underlined. An antisense primer, 5′-GA TCTAGACATATTTAACGACTATCCG-3′, was designed from a bys cDNA and contains an XbaI site (underlined). The sense and antisense primers were used with a bys cDNA in a PCR reaction with Pfu polymerase (Stratagene). The resultant PCR product encodes the C-terminus of the myc epitope fused in-frame to the Bys coding sequence. In a series of subcloning steps, a 159-bp HindIII to ClaI fragment of the PCR product was joined to the HindIII site in a myc epitope (taken from a plasmid with myc-tagged rat S6KI, kindly provided by George Thomas) and to the ClaI site of a bys cDNA that had not been subjected to PCR. This generated a plasmid with the full myc epitope fused in-frame to the coding region of the bys cDNA. The entire construct was sequenced on both DNA strands and then cloned into the expression vector pPAAT under the control of an α-tubulin promoter. pPAAT was made by removing an EcoRI to KpnI fragment containing an α-tubulin promoter and a BamHI to XbaI fragment containing an SV40 polyadenylation signal from the vector pBD1119 (kindly provided by Barry Dickson) and subcloning these components into pGem3Z (Promega).
Cell culture, transfection and analysis of Bys protein
S2 cells were cultured in M3 media supplemented with 10% insect media supplement and antibiotics (Sigma). Cells were plated in 4 ml media at 2×106 cells/ml in 60-mm culture plates and transfected with 5 μg pPAAT or pPAAT-myc-Bys DNA using DDAB as described previously (Han 1996). For western blot analysis, cells were pipetted from dishes, pelleted by centrifugation at 724×g, washed two times in PBS, lysed in 75 μl 1× SDS sample loading buffer (62.5 mM Tris-HCl [pH 6.8]; 2% SDS; 10% glycerol; 50 mM DTT; 0.01% bromophenol blue) and 30 μl lysates were separated on a 12% SDS-polyacrylamide gel. Proteins were transferred to a PDVF membrane (Pall Biosciences). A mouse monoclonal antibody to the myc epitope was isolated from the supernatant of the cell line 1–9E10.2 (purchased from American Type Culture Collection). The 9E10 antibody and a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (Sigma) were used at 1:5,000 dilutions. Proteins were detected by enhanced chemiluminescence (Amersham). For immunostaining, S2 cells were cultured, transfected, collected by centrifugation at 1,125×g and washed in PBS as described above. Cells were resuspended in PBS and allowed to settle on poly-L-lysine coated slides. Immunostaining was done as described in Orosz et al. (1996) except that 4% formaldehyde was used and, after washing to remove excess secondary antibody, cells were stained with 1 μg/ml DAPI in PBS for 10 s followed by extensive washing in PBS. The 9E10 antibody and a goat anti-mouse FITC-conjugated secondary antibody (Jackson ImmunoResearch) were used at 1:500 dilutions. Cells were examined with a Nikon CARV confocal microscope and images were processed with Adobe PhotoShop.
Results and discussion
Maternal bys expression
Whole-mount in situ hybridization to adult ovaries shows that maternal bys mRNA is present in nurse cells of stage 5–6 egg chambers (Fig. 1a). In stage 10 egg chambers, maternal bys transcripts are clearly visible in nurse cells, but not in the follicle cells overlying the oocyte (Fig. 1b). Maternal bys mRNA is found ubiquitously in preblastoderm embryos (Fig. 1c) and is depleted during cellularization (Fig. 1d).
Spatial expression of bys in ovaries and embryos. Wild-type ovaries (Oregon R) and embryos (w1118) were hybridized in situ with a bys anti-sense RNA probe. Egg chambers at a stage 5–6 and b stage 10. Embryos are at the following stages: c preblastoderm; d late stage 5, cellular blastoderm; e stage 7, early gastrulation; f stage 10, germband extended; g, h stage 13, germband retracted; i stage 16, three gut constrictions. Indicated in f are the anal pads (arrow), clypeolabrum (arrowhead), salivary glands (double arrows), anterior midgut (amg), posterior midgut (pmg) and mesoderm (md). The inset in f shows a control stage 10 embryo hybridized with a sense probe. Embryos are shown laterally except for the embryo shown dorsally in h. All embryos are oriented with the anterior to the left
Embryonic and larval bys expression
In situ hybridization to embryos shows that zygotic bys expression is temporally dynamic and tissue specific. Zygotic bys expression is ubiquitous and relatively weak in gastrulating stage 8 embryos (Fig. 1e), but in germband extended embryos, bys expression is strong and specifically localized (Fig. 1f). bys expression is strong in the anterior and posterior midgut, but low or absent in the foregut and hindgut (Fig. 1f, g). bys is also expressed in the mesodermal component of the germband, the clypeolabrum and the anal pads (Fig. 1f, g). In a dorsal view of stage 13 embryos, bys expression is apparent in the midgut and the underlying visceral mesoderm (Fig. 1h). In later stages, bys expression is restricted primarily to the midgut of embryos (Fig. 1i). These results are similar to those reported by the Berkeley Drosophila Genome Project (FlyBase 1999) but show additional details of bys expression such as in the salivary gland primordia, clypeolabrum and anal pads.
In larval imaginal discs, bys expression is restricted to specific regions. bys is strongly expressed in the dorsal and ventral regions of the wing pouch that will form the epithelia of the adult wing while weaker bys expression can be seen in the notum (Fig. 2a). In leg discs, bys is expressed in central regions (Fig. 2b, d). In haltere discs, bys is expressed in the haltere pouch and in a dorsal and anterior wedge (Fig. 2c).
bys mRNA expression in imaginal discs of third instar larvae. a Wild-type (w1118) wing discs show bys expression in the dorsal (asterisk) and ventral (arrow) wing pouch and in the notum (arrowhead). b, d Leg discs show expression in central regions. c A haltere disc shows expression in the haltere pouch (arrow) and an anterior wedge (arrowhead)
The embryonic and larval bys expression patterns could indicate a role for Bys in adhesion or other cell-type specific events. For example, bys is expressed in cells of the midgut primordia which change shape as they invaginate, undergo an epithelial to mesenchyme transition, migrate over the visceral mesoderm and then convert back to epithelium once they meet in the middle of the embryo to form the midgut (Skaer 1993). bys expression occurs strongly in the region of the wing pouch which gives rise to two epithelial sheets of the adult wing that adhere to one another after the disc everts. Although cytokeratins, with which human Bystin interacts, are present in epithelia of most animals, sequences coding for cytokeratins have not been found in the Drosophila genome (Goldstein and Gunawardena 2000). Thus, if Bys has a role in adhesion in the wing epithelia, it is unlikely to be through association with cytokeratins.
Alternatively, bys mRNA expression patterns could reflect a biosynthetic role. Maternal bys is highly expressed in the nurse cells that carry out high levels of rRNA synthesis and ribosome production (Spradling 1993). Additionally, the embryonic bys mRNA expression pattern is almost identical to those of pitchoune (pit) and modulo (mod), genes that are potential transcriptional targets of Drosophila Myc (dMyc), a known regulator of cell growth. Despite the tissue-specific embryonic expression of pit and mod, these genes both encode nucleolar proteins. Mutations in these genes cause delayed development, growth defects and lethality. In the case of mutation of pit, growth of endoreplicative and mitotic cells is impacted while mutation of mod has effects primarily on growth of proliferating cells (Zaffron et al. 1998; Perrin et al. 2003).
Epitope-tagged Bys is expressed as a 60-kDa protein and is nuclear in Schneider line 2 cells
To ask if properties of the Bys protein would provide insight into potential Bys function, we generated a cDNA coding for a myc epitope fused to the N-terminus of Bys (termed myc-Bys) and transiently transfected this construct into Drosophila S2 cells. We first asked if myc-Bys is potentially modified post-translationally by examining its migration by SDS-PAGE and carrying out western blot analysis with the 9E10 antibody. myc-Bys migrates as a single band with an apparent molecular weight (MW) of 60 kDa (Fig. 3). Conceptual translation of the bys cDNA shows that the Bys protein consists of 436 amino acids with a calculated MW of 49.97 kDa. The calculated MW of myc-Bys is 51.16 kDa. Analysis of Bys with PROSITE (Swiss Institute of Bioinformatics; Gasteiger et al. 2003) reveals that Bys has potential phosphorylation sites for protein kinase C, casein kinase II, tyrosine kinases and cAMP and cGMP dependent kinases. All of these phosphorylation sites have a high probability of occurrence and further work will be necessary to determine if Bys is phosphorylated. A myc-tagged yeast Enp1 protein was observed to migrate on SDS-PAGE at an apparent MW of 70 kDa, larger than the predicted MW of 57.4 kDa. This aberrant migration was not caused by N-linked glycosylation and it was suggested that an acidic domain within Enp1 may cause aberrant mobility (Roos et al. 1997). This acidic domain is conserved in Drosophila Bys and may account for the migration of myc-Bys.
Detection of myc-Bys in Schneider line 2 (S2) cells. Western blot of extracts from S2 cells transiently transfected with myc-Bys (lane 1) or vector alone (lane 2). A single band corresponding to myc-Bys has an apparent MW of 60 kDa. Endogenous cellular proteins of ~20 kDa that cross-react with the 9E10 antibody are faintly visible in both lanes. Longer exposure of the film shows two additional cross-reacting bands of less than 20 kDa in both lanes (not shown)
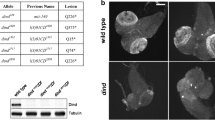
We next used indirect immunofluorescence to examine the sub-cellular location of myc-Bys in S2 cells. This reveals that Bys is localized to the nucleus (Fig. 4b, d). Detection of Bys in the nucleus is specific since the FITC signal is not detectable in all cells (Fig. 4b) or in control cells transfected with vector alone and incubated with the primary and secondary antibodies (not shown). Examining Bys with PredictNLS (Nair et al. 2003) reveals that Bys contains a predicted nuclear localization signal, KKKSH, at amino acids 387–391. Further work will be necessary to determine if this sequence is responsible for nuclear targeting.
Sub-cellular localization of myc-Bys in Schneider line 2 (S2) cells. a S2 cells are shown with differential interference contrast (DIC). b The myc-Bys protein is visualized by FITC (green) in transfected cells. Comparing panels a and b shows that non-transfected cells do not show a FITC signal. c Nuclei were detected with DAPI (blue). d An overlay of the DIC and FITC images. The scale bar in a =20 μm
The proposed functions of the Bys-like proteins in yeast and in human are quite distinct. Our results show that bys mRNA is expressed in a dynamic pattern during development that could be consistent with either a role in cell-cell interactions or in cell growth. However, the observation that embryonic bys expression mirrors that of two dMyc targets that encode nucleolar proteins is intriguing, especially in light of the fact that the 5′ region of the bys gene contains a canonical E-box binding site for Myc. The observation that myc-Bys is found in the nucleus is consistent with the hypothesis that Bys function may be analogous to that of yeast Enp1. Additional work will be required to test this hypothesis. An antibody to the Drosophila Bys protein would be useful for determining whether endogenous Bys is nuclear or cytoplasmic in tissues such as the imaginal discs where bys mRNA expression is spatially restricted. Additionally, RNA interference and the analysis of bys mutant phenotypes should be useful for determining Bys function in Drosophila. Further study of Bys-like proteins in Drosophila and other organisms should reveal if the conserved bys-like family of genes has conserved or distinct functions within phylogenetic groups.
References
Aoki R, Fukuda MN (2000) Recent molecular approaches to elucidate the mechanism of embryo implantation: trophinin, bystin, and tastin as molecules involved in the initial attachment of blastocysts to the uterus in humans. Semin Reprod Med 18:265–271
Chen W, Bucaria J, Band DA, Sutton A, Sternglanz R (2003) Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res 31:690–699
Deng W-M, Leaper K, Bownes M (1999) A targeted gene silencing technique shows that Drosophila myosin VI is required for egg chamber and imaginal disc morphogenesis. J Cell Sci 112:3677–3690
FlyBase (1999) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res 27:85–88. http://flybase.bio.indiana.edu/
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788
Goldstein SB, Gunawardena S (2000) Flying through the Drosophila cytoskeletal genome. J Cell Biol 150:F63–F68
Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, Gavin AC, Hurt E (2002) 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10:105–115
Han K (1996) An efficient DDAB-mediated transfection of Drosophila S2 cells. Nucleic Acids Res 24:4362–4363
Lehmann R, Tautz D (1994) In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA (eds) Methods in cell biology, vol 44. Academic, New York, pp 575–598
Nadano D, Sugihara K, Paria BC, Saburi S, Copeland NG, Gilbert DJ, Jenkins NA, Nakayama J, Fukuda MN (2002) Significant differences between mouse and human trophinins are revealed by their expression patterns and targeted disruption of mouse trophinin gene. Biol Reprod 66:313–321
Nair R, Carter P, Rost B (2003) NLSdb: database of nuclear localization signals. Nucleic Acids Res 31:397–399
O’Neill JW, Bier E (1994) Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. BioTech 17:870–875
Orosz A, Wisniewski J, Wu C (1996) Regulation of Drosophila heat shock factor trimerization: global sequence requirements and independence of nuclear localization. Mol Cell Biol 16:7018–7030
Perrin L, Benassayag C, Morello D, Pradel J, Montagne J (2003) Modulo is a target of Myc selectively required for growth of proliferative cells in Drosophila. Mech Dev 120:645–655
Roos J, Luz JM, Centoducati S, Sternglanz R, Lennarz WJ (1997) ENP1, an essential gene encoding a nuclear protein that is highly conserved from yeast to humans. Gene 185:137–146
Skaer H (1993) The alimentary canal. In: Bate M, Martinez Arias A (eds) The development of Drosophila melanogaster, vol 2. Cold Spring Harbor Laboratory, Cold Spring Harbor, pp 941–1012
Spradling A (1993) Developmental genetics of oogenesis. In: Bate M, Martinez Arias A (eds) The development of Drosophila melanogaster, vol 2. Cold Spring Harbor Laboratory, Cold Spring Harbor, pp 1–70
Suzuki N, Nakayama J, Shih IM, Aoki D, Nozawa S, Fukuda MN (1999) Expression of trophinin, tastin, and bystin by trophoblast and endometrial cells in human placenta. Biol Reprod 60:621–627
Zaffran S, Chartier A, Gallant P, Astier M, Arquier N, Doherty D, Gratecos D, Semeriva M (1998) A Drosophila helicase gene, pitchoune, is required for cell growth and proliferation and is a potential target of d-Myc. Development 125:3571–3584
Acknowledgements
We thank Hélène Barcelo for suggestions and construction of pPAAT, Delcy Rodriguez and Jerzy Bilski for assistance, Jayma Moore and Tom Freeman for assistance with confocal microscopy, and H. Barcelo, Glen McBride, Carrie Leopold and Ron Hutchison for comments on the manuscript. This work was supported by NIH grants GM60956 and GM067756 to M.J.S. E.K.N. was an AURA fellow funded by ND EPSCoR through NSF grant OSR-9452892.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by D.A. Weisblat
Rights and permissions
About this article
Cite this article
Stewart, M.J., Nordquist, E.K. Drosophila Bys is nuclear and shows dynamic tissue-specific expression during development. Dev Genes Evol 215, 97–102 (2005). https://doi.org/10.1007/s00427-004-0447-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00427-004-0447-8