Abstract
Main conclusion
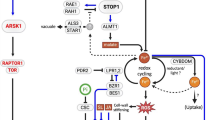
Root transcriptomics and biochemical analyses in water-stressed Pisum sativum plants inoculated with Pseudomonas spp. suggested preservation of ABA-related pathway and ROS detoxification, resulting in an improved tolerance to stress.
Abstract
Drought already affects agriculture in large areas of the globe and, due to climate change, these areas are predicted to become increasingly unsuitable for agriculture. For several years, plant growth-promoting bacteria (PGPB) have been used to improve legume yields, but many aspects of this interaction are still unclear. To elucidate the mechanisms through which root-associated PGPB can promote plant growth in dry environments, we investigated the response of pea plants inoculated with a potentially beneficial Pseudomonas strain (PK6) and subjected to two different water regimes. Combined biometric, biochemical, and root RNA-seq analyses revealed that PK6 improved pea growth specifically under water deficit, as inoculated plants showed an increased biomass, larger leaves, and longer roots. Abscisic acid (ABA) and proline quantification, together with the transcriptome analysis, suggested that PK6-inoculated plant response to water deficit was more diversified compared to non-inoculated plants, involving alternative metabolic pathways for the detoxification of reactive oxygen species (ROS) and the preservation of the ABA stress signaling pathway. We suggest that the metabolic response of PK6-inoculated plants was more effective in their adaptation to water deprivation, leading to their improved biometric traits. Besides confirming the positive role that PGPB can have in the growth of a legume crop under adverse conditions, this study offers novel information on the mechanisms regulating plant–bacteria interaction under varying water availability. These mechanisms and the involved genes could be exploited in the future for the development of legume varieties, which can profitably grow in dry climates.





Similar content being viewed by others
Data availability
The data supporting the findings of this study are available in the Supplementary Information of this article. Reads from RNA-seq were submitted to NCBI Sequence Read Archive (SRA) under Bioproject accession number PRJNA980113.
Abbreviations
- DAT:
-
Days after the transplant
- DEG:
-
Differentially expressed gene
- FC:
-
Fold change
- GO:
-
Gene ontology
- NI:
-
Non-inoculated
- PGPB:
-
Plant growth-promoting bacteria
- POD:
-
Peroxidase
- ROS:
-
Reactive oxygen species
- TRL:
-
Total root length
- WR:
-
Water regime
References
Abd El-Daim IA, Bejai S, Meijer J (2014) Improved heat stress tolerance of wheat seedlings by bacterial seed treatment. Plant Soil 379:337–350. https://doi.org/10.1007/s11104-014-2063-3
Ahsan N, Donnart T, Nouri MZ, Komatsu S (2010) Tissue-specific defense and thermo-adaptive mechanisms of soybean seedlings under heat stress revealed by proteomic approach. J Proteome Res 9(8):4189–4204
Aleem M, Riaz A, Raza Q, Aleem M, Aslam M, Kong K, Atif RM, Kashif M, Bhat JA, Zhao T (2022) Genome-wide characterization and functional analysis of class III peroxidase gene family in soybean reveal regulatory roles of GsPOD40 in drought tolerance. Genomics 114:45–60. https://doi.org/10.1016/j.ygeno.2021.11.016
Álvarez-Aragón R, Palacios JM, Ramírez-Parra E (2023) Rhizobial symbiosis promotes drought tolerance in Vicia sativa and Pisum sativum. Environ Exp Bot 208:105268. https://doi.org/10.1016/j.envexpbot.2023.105268
Alves-Carvalho S, Aubert G, Carrère S, Cruaud C, Brochot AL, Jacquin F, Klein A, Martin C, Boucherot K, Kreplak J, da Silva C, Moreau S, Gamas P, Wincker P, Gouzy J, Burstin J (2015) Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J 84:1–19. https://doi.org/10.1111/tpj.12967
Al-Whaibi MH (2011) Plant heat-shock proteins: a mini review. J King Saud Univ Sci 23:139–150. https://doi.org/10.1016/j.jksus.2010.06.022
Bao Y, Chen C, Fu L, Chen Y (2021) Comparative transcriptome analysis of Rosa chinensis ‘Old Blush’ provides insights into the crucial factors and signaling pathways in salt stress response. Agron J 113:3031–3050. https://doi.org/10.1002/agj2.20715
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207. https://doi.org/10.1007/BF00018060
Belimov AA, Zinovkina NY, Safronova VI, Litvinsky VA, Nosikov VV, Zavalin AA, Tikhonovich IA (2019) Rhizobial ACC deaminase contributes to efficient symbiosis with pea (Pisum sativum L.) under single and combined cadmium and water deficit stress. Environ Exp Bot 167:103859. https://doi.org/10.1016/j.envexpbot.2019.103859
Ben Rejeb K, Abdelly C, Savouré A (2014) How reactive oxygen species and proline face stress together. Plant Physiol Biochem 80:278–284. https://doi.org/10.1016/j.plaphy.2014.04.007
Bendre AD, Ramasamy S, Suresh CG (2018) Analysis of Kunitz inhibitors from plants for comprehensive structural and functional insights. Int J Biol Macromol 113:933–943. https://doi.org/10.1016/j.ijbiomac.2018.02.148
Bharath P, Gahir S, Raghavendra AS (2021) Abscisic acid-induced stomatal closure: an important component of plant defense against abiotic and biotic stress. Front Plant Sci 12:615114. https://doi.org/10.3389/fpls.2021.615114
Brilli F, Pollastri S, Raio A, Baraldi R, Neri L, Bartolini P, Podda A, Loreto F, Maserti BE, Balestrini R (2019) Root colonization by Pseudomonas chlororaphis primes tomato (Lycopersicum esculentum) plants for enhanced tolerance to water stress. J Plant Physiol 232:82–93
Bündig C, Jozefowicz AM, Mock HP, Winkelmann T (2016) Proteomic analysis of two divergently responding potato genotypes (Solanum tuberosum L.) following osmotic stress treatment in vitro. J Proteom 143:227–241
Casanovas EM, Barassi CA, Sueldo RJ (2002) Azospiriflum inoculation mitigates water stress effects in maize seedlings. Cereal Res Commun 30:343–350. https://doi.org/10.1007/BF03543428
Chakraborty K, Jena P, Mondal S, Dash GK, Ray S, Baig MJ, Swain P (2022) Relative contribution of different members of OsDREB gene family to osmotic stress tolerance in indica and japonica ecotypes of rice. Plant Biol 24:356–366. https://doi.org/10.1111/plb.13379
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Deng Y, Lu S (2017) Biosynthesis and regulation of phenylpropanoids in plants. Crit Rev Plant Sci 36:257–290. https://doi.org/10.1080/07352689.2017.1402852
Dossa K, Wei X, Li D, Fonceka D, Zhang Y, Wang L, Yu J, Boshou L, Diouf D, Cissé N, Zhang X (2016) Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol 16:171. https://doi.org/10.1186/s12870-016-0859-4
Dudhate A, Shinde H, Tsugama D, Liu S, Takano T (2018) Transcriptomic analysis reveals the differentially expressed genes and pathways involved in drought tolerance in pearl millet [Pennisetum glaucum (L.) R. Br]. PLoS ONE 13:e0195908. https://doi.org/10.1371/journal.pone.0195908
Fadiji AE, Santoyo G, Yadav AN, Babalola OO (2022) Efforts towards overcoming drought stress in crops: revisiting the mechanisms employed by plant growth-promoting bacteria. Front Microbiol 13:962427. https://doi.org/10.3389/fmicb.2022.962427
FAOSTAT (2021) FAO statistical databases (WWW Document). https://www.fao.org/faostat/en/#data/QCL. Accessed 6.16.22.
Ghosh D, Sen S, Mohapatra S (2018) Drought-mitigating Pseudomonas putida GAP-P45 modulates proline turnover and oxidative status in Arabidopsis thaliana under water stress. Ann Microbiol 68:579–594. https://doi.org/10.1007/s13213-018-1366-7
Giacomelli L, Nanni V, Lenzi L, Zhuang J, Dalla Serra M, Banfield MJ, Town CD, Silverstein KAT, Baraldi E, Moser C (2012) Identification and characterization of the defensin-like gene family of grapevine. Mol Plant Microbe Interact 25:1118–1131. https://doi.org/10.1094/MPMI-12-11-0323
Goh CH, Veliz Vallejos DF, Nicotra AB, Mathesius U (2013) The impact of beneficial plant-associated microbes on plant phenotypic plasticity. J Chem Ecol 39:826–839. https://doi.org/10.1007/s10886-013-0326-8
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916. https://doi.org/10.1111/j.1365-313X.2005.02575.x
Huang S, Zhang H, Purves RW, Bueckert R, Tar’an B, Warkentin TD (2023) Comparative analysis of heat-stress-induced abscisic acid and heat shock protein responses among pea varieties. Crop Sci 63(1):139–150
Hussain RM, Ali M, Feng X, Li X (2017) The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol 17:55. https://doi.org/10.1186/s12870-017-1001-y
Ilangumaran G, Subramanian S, Smith DL (2022) Soybean leaf proteomic profile influenced by rhizobacteria under optimal and salt stress conditions. Front Plant Sci 13:809906. https://doi.org/10.3389/fpls.2022.809906
IPCC (2022) IPCC, 2022: climate change 2022: impacts, adaptation, and vulnerability. Contribution of working group ii to the sixth assessment report of the intergovernmental panel on climate change
Kim YC, Glick BR, Bashan Y, Ryu CM (2012) Enhancement of plant drought tolerance by microbes. In: Aroca R (ed) Plant responses to drought stress: from morphological to molecular features. Springer, Berlin, pp 383–413. https://doi.org/10.1007/978-3-642-32653-0_15
Knopkiewicz M, Wojtaszek P (2019) Validation of reference genes for gene expression analysis using quantitative polymerase chain reaction in pea lines (Pisum sativum) with different lodging susceptibility. Ann Appl Biol 174(1):86–91
Kour D, Rana KL, Yadav N, Yadav AN, Kumar A, Meena VS, Singh B, Chauhan VS, Dhaliwal HS, Saxena AK (2019) Rhizospheric microbiomes: biodiversity, mechanisms of plant growth promotion, and biotechnological applications for sustainable agriculture. In: Kumar A, Meena VS (eds) Plant growth promoting rhizobacteria for agricultural sustainability: from theory to practices. Springer, Singapore, pp 19–65. https://doi.org/10.1007/978-981-13-7553-8_2
Kreplak J, Madoui MA, Cápal P, Novák P, Labadie K, Aubert G, Bayer PE, Gali KK, Syme RA et al (2019) A reference genome for pea provides insight into legume genome evolution. Nat Genet 51:1411–1422. https://doi.org/10.1038/s41588-019-0480-1
Kumar M, Yusuf MA, Yadav P, Narayan S, Kumar M (2019) Overexpression of chickpea defensin gene confers tolerance to water-deficit stress in Arabidopsis thaliana. Front Plant Sci 10:290. https://doi.org/10.3389/fpls.2019.00290
Kuromori T, Seo M, Shinozaki K (2018) ABA transport and plant water stress responses. Trends Plant Sci 23:513–522. https://doi.org/10.1016/j.tplants.2018.04.001
Le DT, Nishiyama R, Watanabe Y, Mochida K, Yamaguchi-Shinozaki K, Shinozaki K, Tran LSP (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res 18:263–276. https://doi.org/10.1093/dnares/dsr015
Li J, Guo X, Zhang M, Wang X, Zhao Y, Yin Z, Zhang Z, Wang Y, Xiong H, Zhang H, Todorovska E, Li Z (2018) OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci 270:131–139. https://doi.org/10.1016/j.plantsci.2018.01.017
Liang X, Zhang L, Natarajan SK, Becker DF (2013) Proline mechanisms of stress survival. Antioxid Redox Signal 19(9):998–1011. https://doi.org/10.1089/ars.2012.5074
López CM, Pineda M, Alamillo JM (2020) Differential regulation of drought responses in two Phaseolus vulgaris genotypes. Plants 9:1815. https://doi.org/10.3390/plants9121815
López-Carbonell M, Gabasa M, Jáuregui O (2009) Enhanced determination of abscisic acid (ABA) and abscisic acid glucose ester (ABA-GE) in Cistus albidus plants by liquid chromatography–mass spectrometry in tandem mode. Plant Physiol Biochem 47:256–261. https://doi.org/10.1016/j.plaphy.2008.12.016
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mellidou I, Ainalidou A, Papadopoulou A, Leontidou K, Genitsaris S, Karagiannis E, Van de Poel B, Karamanoli K (2021) Comparative transcriptomics and metabolomics reveal an intricate priming mechanism involved in PGPR-mediated salt tolerance in tomato. Front Plant Sci 12:713984
Merilo E, Laanemets K, Hu H, Xue S, Jakobson L, Tulva I, Gonzalez-Guzman M, Rodriguez PL, Schroeder JI, Broschè M, Kollist H (2013) PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness-, and CO2-induced stomatal regulation. Plant Physiol 162:1652–1668. https://doi.org/10.1104/pp.113.220608
Nishu SD, No JH, Lee TK (2022) Transcriptional response and plant growth promoting activity of Pseudomonas fluorescens DR397 under drought stress conditions. Microbiol Spectr 10:e00979-e1022. https://doi.org/10.1128/spectrum.00979-22
Okamoto M, Peterson FC, Defries A, Park SY, Endo A, Nambara E, Volkman BF, Cutler SR (2013) Activation of dimeric ABA receptors elicits guard cell closure, ABA-regulated gene expression, and drought tolerance. Proc Natl Acad Sci USA 110:12132–12137. https://doi.org/10.1073/pnas.1305919110
Pandey GK, Grant JJ, Cheong YH, Kim BG, Li L, Luan S (2005) ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol 139:1185–1193. https://doi.org/10.1104/pp.105.066324
Pátková I, Čepl JJ, Rieger T, Blahůšková A, Neubauer Z, Markoš A (2012) Developmental plasticity of bacterial colonies and consortia in germ-free and gnotobiotic settings. BMC Microbiol 12:178. https://doi.org/10.1186/1471-2180-12-178
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. https://doi.org/10.1038/nmeth.4197
Pieterse CMJ, Zamioudis C, Berendsen RL, Weller DM, Van Wees SCM, Bakker PAHM (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375. https://doi.org/10.1146/annurev-phyto-082712-102340
Ramakrishna G, Singh A, Kaur P, Yadav SS, Sharma S, Gaikwad K (2022) Genome wide identification and characterization of small heat shock protein gene family in pigeonpea and their expression profiling during abiotic stress conditions. Int J Biol Macromol 197:88–102
Ramos Solano B, Pereyra de la Iglesia MT, Probanza A, Lucas García JA, Megías M, Gutierrez Mañero FJ (2007) Screening for PGPR to improve growth of Cistus ladanifer seedlings for reforestation of degraded mediterranean ecosystems. In: Velázquez E, Rodríguez-Barrueco C (eds) First International meeting on microbial phosphate solubilization, developments in plant and soil sciences. Springer, Dordrecht, pp 59–68. https://doi.org/10.1007/978-1-4020-5765-6_7
Rasheed A, Barqawi AA, Mahmood A, Nawaz M, Shah AN, Bay DH, Alahdal MA, Hassan MU, Qari SH (2022) CRISPR/Cas9 is a powerful tool for precise genome editing of legume crops: a review. Mol Biol Rep 49(6):5595–5609
Raza A, Mubarik MS, Sharif R, Habib M, Jabeen W, Zhang C, Chen H, Chen Z, Siddique KHM, Zhuang W, Varshney RK (2023) Developing drought-smart, ready-to-grow future crops. Plant Genome 16(1):e20279
Saijo Y, Loo EP, Yasuda S (2018) Pattern recognition receptors and signaling in plant–microbe interactions. Plant J 93:592–613. https://doi.org/10.1111/tpj.13808
Saikia J, Sarma RK, Dhandia R, Yadav A, Bharali R, Gupta VK, Saikia R (2018) Alleviation of drought stress in pulse crops with ACC deaminase producing rhizobacteria isolated from acidic soil of Northeast India. Sci Rep 8:3560. https://doi.org/10.1038/s41598-018-21921-w
Schillaci M, Raio A, Sillo F, Zampieri E, Mahmood S, Anjum M, Khalid A, Centritto M (2022) Pseudomonas and Curtobacterium strains from olive rhizosphere characterized and evaluated for plant growth promoting traits. Plants 11:2245. https://doi.org/10.3390/plants11172245
Seethepalli A, York LM (2021) Rhizo vision explorer—interactive software for generalized root image analysis designed for everyone. AoB Plants 13(6):plab056. https://doi.org/10.5281/zenodo.5121845
Shahzad Z, Ranwez V, Fizames C, Marquès L, Le Martret B, Alassimone J, Godè C, Lacombe E, Castillo T, Saumitou-Laprade P, Berthomieu P, Gosti F (2013) Plant Defensin type 1 (PDF 1): protein promiscuity and expression variation within the Arabidopsis genus shed light on zinc tolerance acquisition in Arabidopsis halleri. New Phytol 200(3):820–833
Shekhawat K, Almeida-Trapp M, García-Ramírez GX, Hirt H (2022) Beat the heat: plant- and microbe-mediated strategies for crop thermotolerance. Trends Plant Sci Special Issue Clim Change Sustai II 27:802–813. https://doi.org/10.1016/j.tplants.2022.02.008
Sirohi P, Yadav BS, Afzal S, Mani A, Singh NK (2020) Identification of drought stress-responsive genes in rice (Oryza sativa) by meta-analysis of microarray data. J Genet 99:35. https://doi.org/10.1007/s12041-020-01195-w
Sohn KH, Lee SC, Jung HW, Hong JK, Hwang BK (2006) Expression and functional roles of the pepper pathogen-induced transcription factor RAV1 in bacterial disease resistance, and drought and salt stress tolerance. Plant Mol Biol 61:897–915. https://doi.org/10.1007/s11103-006-0057-0
Soneson C, Love MI, Robinson MD (2016) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4:1521. https://doi.org/10.12688/f1000research.7563.2
Srivastava R, Kobayashi Y, Koyama H, Sahoo L (2023) Cowpea NAC1/NAC2 transcription factors improve growth and tolerance to drought and heat in transgenic cowpea through combined activation of photosynthetic and antioxidant mechanisms. J Integr Plant Biol 65:25–44. https://doi.org/10.1111/jipb.13365
Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6:e21800. https://doi.org/10.1371/journal.pone.0021800
Tiwari S, Lata C, Chauhan PS, Nautiyal CS (2016) Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol Biochem 99:108–117. https://doi.org/10.1016/j.plaphy.2015.11.001
Truong VK, Chapman J, Cozzolino D (2021) Monitoring the bacterial response to antibiotic and time growth using near-infrared spectroscopy combined with machine learning. Food Anal Methods 14:1394–1401
Türkan İ, Bor M, Özdemir F, Koca H (2005) Differential responses of lipid peroxidation and antioxidants in the leaves of drought-tolerant P. acutifolius Gray and drought-sensitive P. vulgaris L. subjected to polyethylene glycol mediated water stress. Plant Sci 168:223–231. https://doi.org/10.1016/j.plantsci.2004.07.032
Vaishnav A, Choudhary DK (2019) Regulation of drought-responsive gene expression in Glycine max L. Merrill is mediated through Pseudomonas simiae Strain AU. J Plant Growth Regul 38:333–342. https://doi.org/10.1007/s00344-018-9846-3
Vurukonda SSKP, Vardharajula S, Shrivastava M, Sk ZA (2016) Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol Res 184:13–24. https://doi.org/10.1016/j.micres.2015.12.003
Wu H, Si Q, Liu J, Yang L, Zhang S, Xu J (2022) Regulation of Arabidopsis matrix metalloproteinases by mitogen-activated protein kinases and their function in leaf senescence. Front Plant Sci 13:864986. https://doi.org/10.3389/fpls.2022.864986
Yang J, Wang H, Zhao S, Liu X, Zhang X, Wu W, Li C (2020) Overexpression levels of LbDREB6 differentially affect growth, drought, and disease tolerance in poplar. Front Plant Sci 11:528550. https://doi.org/10.3389/fpls.2020.528550
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134. https://doi.org/10.1186/1471-2105-13-134
Yıldız M, Terzi H (2021) Comparative analysis of salt-induced changes in the root physiology and proteome of the xero-halophyte Salsola crassa. Braz J Bot 44(1):33–42
Yin H, Li M, Li D, Khan SA, Hepworth SR, Wang SM (2019) Transcriptome analysis reveals regulatory framework for salt and osmotic tolerance in a succulent xerophyte. BMC Plant Biol 19:88. https://doi.org/10.1186/s12870-019-1686-1
Yu X, James AT, Yang A, Jones A, Mendoza-Porras O, Bétrix CA, Ma H, Colgrave ML (2016) A comparative proteomic study of drought-tolerant and drought-sensitive soybean seedlings under drought stress. Crop Pasture Sci 67(5):528–540
Yuwono T, Handayani D, Soedarsono J (2005) The role of osmotolerant rhizobacteria in rice growth under different drought conditions. Aust J Agric Res 56:715–721. https://doi.org/10.1071/AR04082
Zampieri E, Pantelides IS, Balestrini R (2022) Biofertilizers: assessing the effects of plant growth-promoting bacteria (PGPB) or rhizobacteria (PGPR) on soil and plant health. In: Horwath WR (ed) Improving soil health. Burleigh Dodds Science Publishing, Cambridge, pp 1–27. https://doi.org/10.19103/AS.2021.0094.18
Zboralski A, Filion M (2023) Pseudomonas spp. can help plants face climate change. Front Microbiol 14:1198131. https://doi.org/10.3389/fmicb.2023.1198131
Zdunek-Zastocka E, Sobczak M (2013) Expression of Pisum sativum PsAO3 gene, which encodes an aldehyde oxidase utilizing abscisic aldehyde, is induced under progressively but not rapidly imposed drought stress. Plant Physiol Biochem 71:57–66. https://doi.org/10.1016/j.plaphy.2013.06.027
Zhao B, Zhang S, Yang W, Li B, Lan C, Zhang J, Yuan L, Wang Y, Xie Q, Han J, Mur LAJ, Hao X, Roberts JA, Miao Y, Yu K, Zhang X (2021) Multi-omic dissection of the drought resistance traits of soybean landrace LX. Plant Cell Environ 44:1379–1398. https://doi.org/10.1111/pce.14025
Zipfel C, Oldroyd GED (2017) Plant signalling in symbiosis and immunity. Nature 543:328–336. https://doi.org/10.1038/nature22009
Acknowledgements
The authors are grateful to Dr. Raffaella Balestrini for the useful comments and discussion on the study, as well as for critically reviewing the manuscript.
Funding
The work was supported by the National Research Council of Italy — PMAS Arid Agriculture University Rawalpindi-Pakistan Joint Laboratory Project 2021–2023 “Selection and evaluation of plant growth promoting rhizobacteria to increase climate-resilient crop production” (PI: Dr. Mauro Centritto).
Author information
Authors and Affiliations
Contributions
Conceptualization: FS; methodology: MS, FS, EZ, CB, and AG; formal analysis and investigation: MS, FS, and EZ; writing—original draft preparation: MS and EZ; writing—review and editing: MS, FS, and EZ;
Corresponding author
Ethics declarations
Conflict of interest
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Communicated by Stefan de Folter.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1. Fig. S1
: Shoot length of pea plants recorded from 12 to 33 days after the transplant (DAT). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. Symbols represent the mean shoot length of each treatment; n = 5 for NIWR1, NIWR2, n = 4 for PK6WR1 and PK6WR2. (JPG 837 kb)
Supplementary file 2. Fig. S2
: Leaf number and % of wilted leaves. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2; n = 5 for NIWR1 and NIWR2, n = 4 for PK6WR1 and PK6WR2. Individual standard deviations are used to calculate the intervals. Bars represent the standard error of the mean. Above the intervals, different letters display statistical differences among the treatments (Fisher LSD pairwise comparison, 95% confidence). (JPG 120 kb)
Supplementary file 3. Fig. S3
: Concentration (%) of C and N in dry shoots of pea. NI, non-inoculated; WR1, water regime 1; WR2, water regime 2; DW, dry weight; n = 5 for NIWR1 and NIWR2, n = 4 for PK6WR1 and PK6WR2. Individual standard deviations are used to calculate the intervals. Bars represent the standard error of the mean. Above the intervals, different letters display statistical differences among the treatments (Fisher LSD pairwise comparison, 95% confidence). (JPG 111 kb)
Supplementary file 4. Fig. S4
: Correlation plot between expression data obtained by RNA-seq (log2FC by RNA-seq) and RT-qPCR analysis (log2FC by RT-qPCR). Regression line (dark grey line), R-square and associated P-value are shown. The gray shadow represents the 95% confidence interval. Tested genes are labelled as in Table S5. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (JPG 1297 kb)
Supplementary file 5. Fig. S5
: Heatmap displaying the relative expression values (log2FC, comparison between WR2 and WR1) of genes involved in the GO names “proline metabolic process” and “hydrogen peroxide catabolic process”. Genes in white were not significantly up- or down-regulated (Wald test, P-value corrected for multiple testing > 0.05). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (JPG 755 kb)
Supplementary file 6. Fig. S6
: Heatmap displaying the relative expression values (log2FC, comparison between WR2 and WR1) of genes involved in the GO names “response to heat”. Genes in white were not significantly up- or down-regulated (Wald test, P-value corrected for multiple testing > 0.05). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (JPG 579 kb)
Supplementary file 7. Fig. S7
: Heatmap displaying the relative expression values (log2FC, comparison between WR2 and WR1) of genes involved in the GO names “coumarin metabolic process” and “phenylpropanoid metabolic process”. Genes in white were not significantly up- or down-regulated (Wald test, P-value corrected for multiple testing > 0.05). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (JPG 720 kb)
Supplementary file 8. Fig. S8
: Heatmap displaying the relative expression values (log2FC, comparison between WR2 and WR1) of genes involved in the GO names “biological process involved in interspecies interaction between organisms” and “response to fungus”. Genes in white were not significantly up- or down-regulated (Wald test, P-value corrected for multiple testing > 0.05). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (JPG 1333 kb)
Supplementary file 9. Fig. S9
: Heatmap displaying the relative expression values (log2FC, comparison between WR2 and WR1) of genes involved in the GO names “regulation of root hair elongation” and “regulation of leaf senescence”. Genes in white were not significantly up- or down-regulated (Wald test, P-value corrected for multiple testing > 0.05). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (JPG 122 kb)
Supplementary file 10. Fig. S10
: Heatmap displaying the relative expression values (log2FC, comparison between WR2 and WR1) of genes putatively involved in ABA metabolism. Genes in white were not significantly up- or down-regulated (Wald test, P-value corrected for multiple testing > 0.05). (JPG 2022 kb)
Supplementary file 11. Table S1
: Characteristics of primers used in the RT-qPCR analysis on selected DEGs. Transcript sequence IDs are from P. sativum Ensembl database. (XLSX 7 kb)
Supplementary file 12. Table S2
: Summary Statistics of RNA-Seq alignment of pea roots reads. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (XLSX 6 kb)
Supplementary file 13. Table S3
: Raw RNA-seq pea root reads count (TPM). NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (XLSX 3892 kb)
Supplementary file 14. Table S4
: Significant (P adj < 0.05) DEGs obtained comparing the NIWR1, NIWR2, PK6WR1 and PK6WR2 treatments. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (XLSX 691 kb)
Supplementary file 15. Table S5
: RT-qPCR gene expression data of five up-regulated DEGs, five down-regulated DEGs and three DEGs putatively coding for peroxidases obtained in the comparison between WR2 and WR1 plants. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (XLSX 6 kb)
Supplementary file 16. Table S6
: Fold changes of DEGs assigned to GO enriched terms. DEGs in blue font are from the comparison NIWR2 versus NIWR1, DEGs in red font are from the comparison PK6WR2 versus PK6WR1. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (XLSX 126 kb)
Supplementary file 17. Table S7
: Fold changes of genes related to ABA metabolism in the comparisons NIWR2 versus NIWR1 and PK6WR2 versus PK6WR1. NI, non-inoculated; PK6, PK6-inoculated; WR1, water regime 1; WR2, water regime 2. (XLSX 12 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Schillaci, M., Zampieri, E., Brunetti, C. et al. Root transcriptomic provides insights on molecular mechanisms involved in the tolerance to water deficit in Pisum sativum inoculated with Pseudomonas sp.. Planta 259, 33 (2024). https://doi.org/10.1007/s00425-023-04310-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-023-04310-0