Abstract
Main conclusion
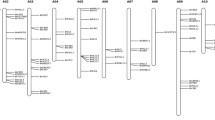
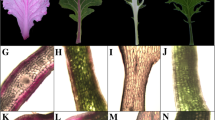
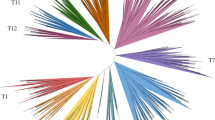
From Brassica oleracea genome, 88 anthocyanin biosynthetic genes were identified. They expanded via whole-genome or tandem duplication and showed significant expression differentiation. Functional characterization revealed BoMYB113.1 as positive and BoMYBL2.1 as negative regulators responsible for anthocyanin accumulation.
Abstract
Brassica oleracea produces various health-promoting phytochemicals, including glucosinolates, carotenoids, and vitamins. Despite the anthocyanin biosynthetic pathways in the model plant Arabidopsis thaliana being well characterized, little is known about the genetic basis of anthocyanin biosynthesis in B. oleracea. In this study, we identified 88 B. oleracea anthocyanin biosynthetic genes (BoABGs) representing homologs of 46 Arabidopsis anthocyanin biosynthetic genes (AtABGs). Most anthocyanin biosynthetic genes, having expanded via whole-genome duplication and tandem duplication, retained more than one copy in B. oleracea. Expression analysis revealed diverse expression patterns of BoABGs in different tissues, and BoABG duplications showed significant expression differentiation. Additional expression analysis and functional characterization revealed that the positive regulator BoMYB113.1 and negative regulator BoMYBL2.1 may be key genes responsible for anthocyanin accumulation in red cabbage and ornamental kale by upregulating the expression of structural genes. This study paves the way for a better understanding of anthocyanin biosynthetic genes in B. oleracea and should promote breeding for anthocyanin content.








Similar content being viewed by others
Data availability
The data generated or used in this study are included in this article and its supplementary materials. RNA-Seq data were obtained from the Gene Expression Omnibus (GEO) database with the accession number GSE42891. BoABGs sequences were retrieved from Bolbase (http://www.ocri-genomics.org/bolbase/index.html). Arabidopsis ABGs sequences were retrieved from Col-0 Arabidopsis reference genome in TAIR (http://www.arabidopsis.org/).
Abbreviations
- ABGs:
-
Anthocyanin biosynthetic genes
- ROS:
-
Reactive oxygen species
- PAL:
-
Phenylalanine ammonia lyase
- C4H:
-
Cinnamate 4-hydroxylase
- 4CL:
-
4-Coumarate: CoA ligase
- CHS:
-
Chalcone synthase
- F3H:
-
Flavanone 3-hydroxylase
- F3′H:
-
Flavonoid 3′-hydroxylase
- FLS:
-
Flavonols by flavonol synthase
- DFR:
-
Dihydroflavonol 4-reductase
- ANS:
-
Anthocyanidin synthase
- GEO:
-
Gene expression omnibus
- WGT:
-
Whole-genome triplication
References
Alamery S, Tirnaz S, Bayer P, Tollenaere R, Chaloub B, Edwards D, Batley J (2018) Genome-wide identification and comparative analysis of NBS-LRR resistance genes in Brassica napus. Crop Pasture Sci 69:72–93
Cheng F, Wu J, Fang L, Sun S, Liu B, Lin K, Bonnema G, Wang X (2012a) Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS One 7:e36442
Cheng F, Wu J, Fang L, Wang X (2012b) Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front Plant Sci 3:198
Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L (2010) The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol 154:1470–1480
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743
Freeling M, Thomas BC (2006) Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res 16:805–814
Ganko EW, Meyers BC, Vision TJ (2007) Divergence in expression between duplicated genes in Arabidopsis. Mol Biol Evol 24:2298–2309
Gould KS (2004) Nature’s swiss army knife: the diverse protective roles of anthocyanins in leaves. J Biomed Biotechnol 2004:314–320
Han F, Zhang X, Yang L, Zhuang M, Zhang Y, Li Z, Fang Z, Lv H (2018) iTRAQ-based proteomic analysis of Ogura-CMS cabbage and its maintainer line. Int J Mol Sci 19(10):3180
Han F, Cui H, Zhang B, Liu X, Yang L, Zhuang M, Lv H, Li Z, Wang Y, Fang Z, Song J, Zhang Y (2019) Map-based cloning and characterization of BoCCD4, a gene responsible for white/yellow petal color in B. oleracea. BMC Genom 20:242
Harborne JB, Williams CA (2001) Anthocyanins and other flavonoids. Nat Prod Rep 18:310–333
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1071
Landi M, Guidi L, Pardossi A, Tattini M, Gould KS (2014) Photoprotection by foliar anthocyanins mitigates effects of boron toxicity in sweet basil (Ocimum basilicum). Planta 240:941–953
Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, Caboche M (2006) Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol 57:405–430
Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5:3930
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Nhukarume L, Chikwambi Z, Muchuweti M, Chipurura B (2010) Phenolic content and antioxidant capacities of Parinari curatelifolia, Strychnos spinosa and Adansonia digitata. J Food Biochem 34:207–221
Parkin IAP, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15:R77
Scalzo RL, Genna A, Branca F, Chedin M, Chassaigne H (2008) Anthocyanin composition of cauliflower (Brassica oleracea L. var. botrytis) and cabbage (B. oleracea L. var. capitata) and its stability in relation to thermal treatments. Food Chem 107:136–144
Sémon M, Wolfe KH (2007) Consequences of genome duplication. Curr Opin Genet Dev 17:505–512
Shi MZ, Xie DY (2014) Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat Biotech 8:47–60
Solovchenko A, Schmitz-Eiberger M (2003) Significance of skin flavonoids for UV-B-protection in apple fruits. J Exp Bot 54:1977–1984
Song H, Yi H, Lee M, Han CT, Lee J, Kim H, Park J, Nou I, Kim S, Hur Y (2018) Purple Brassica oleracea var. capitata F. rubra is due to the loss of BoMYBL2-1 expression. BMC Plant Biol 18:82
Springob K, Nakajima J, Yamazaki M, Saito K (2003) Recent advances in the biosynthesis and accumulation of anthocyanins. Nat Prod Rep 20:288–303
Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B (2010) Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 188:985–1000
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang X, Wang H, Wang J, Sun R, Wu J, Liu S et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43(10):1035
Wang Y, Wang Y, Song Z, Zhang H (2016) Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol Plant 9(10):1395–1405
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223
Xiao Z, Xing M, Liu X, Fang Z, Yang L, Zhang Y, Wang Y, Zhuang M, Lv H (2020) An efficient virus-induced gene silencing (VIGS) system for functional genomics in Brassicas using a cabbage leaf curl virus (CaLCuV)-based vector. Planta 252(3):42
Yan C, An G, Zhu T, Zhang W, Zhang L, Peng L, Chen J, Kuang H (2019) Independent activation of the BoMYB2 gene leading to purple traits in Brassica oleracea. Theor Appl Genet 132:895–906
Zhang Y, Butelli E, Martin C (2014) Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol 19:81–90
Zhao J, Dixon RA (2010) The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci 15:72–80
Zhao L, Gao L, Wang H, Chen X, Wang Y, Yang H, Wei C, Wan X, Xia T (2013) The R2R3-MYB, bHLH, WD40, and related transcription factors in flavonoid biosynthesis. Funct Integr Genom 3:75–98
Acknowledgements
This work was supported by grants from the State Key Laboratory of Vegetable Germplasm Innovation (201902), the Major State Research Development Program (2016YFD0101702), the National Science Foundation of China (31572141), the Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences (CAAS-ASTIP-IVFCAAS), and the earmarked fund for the Modern Agro-Industry Technology Research System, China (nycytx-35-gw01). The work reported herein was performed in the Key Laboratory of Biology and Genetic Improvement of Horticultural Crops, Ministry of Agriculture, Beijing 100081, China. These funding bodies had no role in the design of the study; collection, analysis, or interpretation of the data; or writing of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Additional information
Communicated by Anastasios Melis.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2021_3746_MOESM1_ESM.xlsx
Supplementary Table S1: The A. thaliana anthocyanin pathway genes their B. oleracea orthologs and detailed information of BoABGs. (XLSX 69 KB)
Rights and permissions
About this article
Cite this article
Han, F., Zhang, X., Yang, L. et al. Genome-wide characterization and analysis of the anthocyanin biosynthetic genes in Brassica oleracea. Planta 254, 92 (2021). https://doi.org/10.1007/s00425-021-03746-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03746-6