Abstract
Main conclusion
A total of 249 sites from 197 proteins showed a differential ubiquitination level in the fiber development of ramie barks. The function of two differentially ubiquitinated proteins for fiber growth was demonstrated.
Abstract
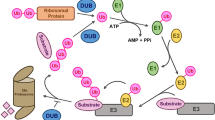
Ubiquitination is one of the most common post-translational modifications of proteins, and it plays essential roles in plant growth and development. However, the involvement of ubiquitination in the growth of plant fibers remains largely unknown. We compared the ubiquitylome of the top and middle stems of ramie bark, with different fiber growth stages. We identified 249 differentially ubiquitinated sites in 197 proteins in fiber-developing barks in the stems and found that seven were homologs of Arabidopsis proteins associated with fiber growth. Overexpression of the differentially ubiquitinated proteins, RWA3 homolog whole_GLEAN_10024150 and MYB protein whole_GLEAN_10015497, significantly promoted fiber growth in transgenic Arabidopsis, indicating their involvement in this process. We also found that the abundance of these proteins decreased when their ubiquitination levels increased and vice versa in the fiber-developing bark. These results indicated that the abundance of these two proteins was adjusted through ubiquitin-dependent degradation. Collectively, our findings provide important insights into the involvement of ubiquitination in the growth of ramie fibers.



Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the data set identifier PXD024923.
Abbreviations
- GO:
-
Gene Ontology
- MPS:
-
Bast in middle part of stem
- PTM:
-
Post-translational modification
- TPS:
-
Bast in top part of stem
References
Aldaba VC (1927) The structure and development of the cell wall in plants I. Bast fibers of Boehmeria and Linum. Amer J Bot 14(1):16–24
Beltrao P, Bork P, Krogan NJ, Noort V (2013) Evolution and functional crosstalk of protein post-translational modifications. Mol Syst Biol 9:1–13
Brill E, Van Thournout M, White RG, Llewellyn D, Campbell PM, Engelen S, Ruan Y, Arioli T, Furbank RT (2011) A novel isoform of sucrose synthase is targeted to the cell wall during secondary cell wall synthesis in cotton fiber. Plant Physiol 157(1):40–54
Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua N (2007) The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19(9):2952–2966
Chen J, Pei Z, Dai L, Wang B, Liu L, An X, Peng D (2014) Transcriptome profiling using pyrosequencing shows genes associated with bast fiber development in ramie (Boehmeria nivea L.). BMC Genomics 15(1):919
Chen J, Rao J, Wang Y, Zeng Z, Liu F, Tang Y, Chen X, Liu C, Liu T (2019) Integration of quantitative trait loci mapping and expression profiling analysis to identify genes potentially involved in ramie fiber lignin biosynthesis. Genes 10:842
Feng H, Li X, Chen H, Deng J, Zhang C, Liu J, Wang T, Zhang X, Dong J (2018) GhHUB2, a ubiquitin ligase, is involved in cotton fiber development via the ubiquitin–26S proteasome pathway. J Exp Bot 69:5059–5075
Friso G, Van Wijk KJ (2015) Update: post-translational protein modifications in plant metabolism. Plant Physiol 169(3):1469–1487
Horton P, Park K, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587
Jensen ON (2004) Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol 8(1):33–41
Lee C, Teng Q, Zhong R, Ye ZH (2011) The four Arabidopsis REDUCED WALL ACETYLATION genes are expressed in secondary wall-containing cells and required for the acetylation of xylan. Plant Cell Physiol 52:1289–1301
Li HL (1970) The origin of cultivated plants in Southeast Asia. Econ Bot 24(1):3–19
Li F, Wang Y, Yan L, Zhu S, Liu T (2021) Characterization of the expression profiling of circRNAs in the barks of stems in ramie. Acta Agron Sin 47:1020–1030
Liu T, Zhu S, Tang Q, Chen P, Yu Y, Tang S (2013) De novo assembly and characterization of transcriptome using Illumina paired-end sequencing and identification of CesA gene in ramie (Boehmeria nivea L. Gaud). BMC Genomics 14:125
Liu T, Zhu S, Tang Q, Tang S (2014a) Identification of 32 full-length NAC transcription factors in ramie (Boehmeria nivea L. Gaud) and characterization of the expression pattern of these genes. Mol Genet Genomics 289(4):675–684
Liu T, Zhu S, Tang Q, Tang S (2014b) QTL mapping for fiber yield-related traits by constructing the first genetic linkage map in ramie (Boehmeria nivea L. Gaud). Mol Breeding 34(3):883–892
Liu C, Zhu S, Tang S, Wang H, Zheng X, Chen X, Dai Q, Liu T (2017) QTL analysis of four main stem bark traits using a GBS-SNP-based high-density genetic map in ramie. Sci Rep 7(1):13458
Liu C, Yu H, Li L (2019) SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet 15:e1007928
Luan M, Jian J, Chen P, Chen J, Chen J, Gao Q, Gao G, Zhou JH, Chen K, Guang X et al (2018) Draft genome sequence of ramie, Boehmeria nivea (L.) Gaudich. Mol Ecol Resour 18(3):639–645
Manabe Y, Verhertbruggen Y, Gille S, Harholt J, Chong S, Pawar P, Mellerowicz E, Tenkanen M, Cheng K, Pauly M, Scheller H (2013) Reduced wall acetylation proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol 163(3):1107–1117
Mazzucotelli E, Mastrangelo AM, Crosatti C, Guerra D, Stanca AM, Cattivelli L (2008) Abiotic stress response in plants: when post-transcriptional and post-translational regulations control transcription. Plant Sci 174(4):420–431
Miura K, Hasegawa PM (2010) Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol 20(4):223–232
Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6:288
Nota B (2017) Gogadget: An R package for interpretation and visualization of GO enrichment results. Mol Inf 36:1600132
Raes J, Rohde A, Christensen JH, Peer YV, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133(3):1051–1071
Sen T, Reddy HN (2011) Various industrial applications of hemp, kenaf, flax and ramie natural fibres. IJIMT 2:192–198
Spoel SH (2018) Orchestrating the proteome with post-translational modifications. J Exp Bot 19:4499–4503
Stulemeijer I, Joosten M (2008) Post-translational modification of host proteins in pathogen-triggered defence signalling in plants. Mol Plant Pathol 9(4):545–560
Sulis DB, Wang JP (2020) Regulation of lignin biosynthesis by post-translational protein modifications. Front Plant Sci 11:914
Tang Y, Liu F, Xing H, Mao K, Chen G, Guo Q, Chen J (2019) Correlation analysis of lignin accumulation and expression of key genes involved in lignin biosynthesis of ramie (Boehmeria nivea). Genes 10(5):389
Taylor NG, Gardiner JC, Whiteman R, Turner SR (2004) Cellulose synthesis in the Arabidopsis secondary cell wall. Cellulose 11(3–4):329–338
Tian ZJ, Yi R, Chen JR, Guo QQ, Zhang XW (2008) Cloning and expression of cellulose synthase gene in ramie [Boehmeria nivea (Linn.) Gaud.]. Acta Agron Sin 34:76–83
Tyanova S, Temu T, Cox J (2016) The MaxQuant computational platform for mass spectrometry–based shotgun proteomics. Nat Protoc 11(12):2301–2319
Wang J, Huang JS, Hao XY, Feng YP, Cai YJ, Sun LQ (2014) miRNAs expression profile in bast of ramie elongation phase and cell wall thickening and end wall dissolving phase. Mol Biol Reports 41:901–907
Wang JP, Chuang L, Loziuk PL, Chen H, Lin YC, Shi R, Qu G, Muddiman D, Sederoff R, Chiang V (2015) Phosphorylation is an on/off switch for 5-hydroxyconiferaldehyde O-methyltransferase activity in poplar monolignol biosynthesis. Proc Natl Acad Sci USA 112(27):8481–8486
Wang Z, Yang Z, Li F (2019) Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol J 17(9):1706–1722
Wang Y, Li F, He Q, Bao Z, Zeng Z, An D, Zhang T, Yan L, Wang H, Zhu S, Liu T (2021) Genomic analyses provide comprehensive insights into the domestication of bast fiber crop ramie (Boehmeria nivea). Plant J. https://doi.org/10.1111/tpj.15346
Wei Z, Qu Z, Zhang L, Zhao S, Bi Z, Ji X, Wang X, Wei H (2015) Overexpression of poplar xylem sucrose synthase in tobacco leads to a thickened cell wall and increased height. PLoS ONE 10(3):e0120669
Yamaguchi M, Kubo M, Fukuda H, Demura T (2008) VASCULAR-RELATED NAC-DOMAIN7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J 55(4):652–664
Ye J, Zhang Z, Long H, Zhang Z, Hong Y, Zhang X, You C, Liang W, Ma H, Lu P (2015) Proteomic and phosphoproteomic analyses reveal extensive phosphorylation of regulatory proteins in developing rice anthers. Plant J 84(3):527–544
Zeng Z, Wang Y, Liu C, Yang X, Wang H, Li F, Liu T (2019) Linkage mapping of quantitative trait loci for fiber yield and its related traits in the population derived from cultivated ramie and wild B. nivea var. tenacissima. Sci Rep 9:16855
Zhang M, Leong HW (2010) Bidirectional best hit r-window gene clusters. BMC Bioinformatics 11:S63
Zhang X, Henriques R, Lin SS, Niu QW, Chua NH (2006) Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc 1(2):641
Zhang X, Gou M, Liu CJ (2013) Arabidopsis Kelch repeat F-Box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25(12):4994–5010
Zheng L, Chen Y, Ding D, Zhou Y, Ding L, Wei J, Wang H (2019) Endoplasmic reticulum–localized UBC34 interaction with lignin repressors MYB221 and MYB156 regulates the transactivity of the transcription factors in Populus tomentosa. BMC Plant Biol 19:97
Zhong R, Ye ZH (2015a) Secondary cell walls: biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56:195–214
Zhong R, Ye ZH (2015b) Molecular control of wood formation in trees. J Exp Bot 66:4119–4131
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31871678, 32001512), the Agricultural Science and Technology Innovation Program of China (CAAS-ASTIP-IBFC), and the National Modern Agroindustry Technology Research System of China (nycytx-19-E16).
Author information
Authors and Affiliations
Contributions
HQ analyzed the ubiquitylome. ZZ performed the overexpression analysis of two genes. LF collected tissue samples and extracted proteins. HR conducted the bioinformatic analysis. WY managed the project and contributed novel reagents. LT designed this study and wrote the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Conflicts of interest
The authors have no conflicts of interest to declare.
Additional information
Communicated by Dorothea Bartels.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
425_2021_3652_MOESM1_ESM.docx
Supplementary file 1 (DOCX 1244 KB) Table S1. Primer sequences for DNA amplification. Fig. S1. Microscopy findings of fiber cells from ramie stem barks (from Chen et al. 2014, BMC Genomics 15:919). We analyzed ubiquitylomes in bark from top (TPS) and from middle (MPS) portions of stems, where growth of secondary cellular walls had, respectively, not started and had thickened. Red arrows indicate the differential thickness in the cell wall of fibers from two tissues. Bars in the microscopic section of TPS and MPS is 20 μm long, and in the figure of whole-plant plant is 10 cm long. Fig. S2. Evaluation of repeatability among replicates based on principal component correlation analysis. Blue and red dots, replicate bark samples from top (TPS) and middle (MPS) parts of stems. Fig. S3. Expression of seven fiber growth-related candidates in the bark from the top part of the stem (TPS) and the middle part of the stem (MPS), where the fiber growth is under different stage. The y-axis represents the value of fragments per kilobase per million reads (FPKM).
Rights and permissions
About this article
Cite this article
He, Q., Zeng, Z., Li, F. et al. Ubiquitylome analysis reveals the involvement of ubiquitination in the bast fiber growth of ramie. Planta 254, 1 (2021). https://doi.org/10.1007/s00425-021-03652-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00425-021-03652-x