Abstract
Main Conclusion
Specific sequences within the leader intron of a soybean polyubiquitin gene stimulated gene expression when placed either within a synthetic intron or upstream of a core promoter.
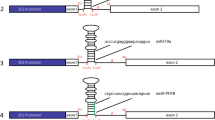
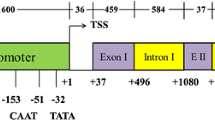
The intron in the 5′ untranslated region of the soybean polyubiquitin promoter, Gmubi, seems to contribute to the high activity of this promoter. To identify the stimulatory sequences within the intron, ten different sequential intronic sequences of 40 nt were isolated, cloned as tetrameric repeats and placed upstream of a minimal cauliflower mosaic virus 35S (35S) core promoter, which was used to control expression of the green fluorescent protein. Intron fragment tetramers were also cloned within a modified, native intron, creating a Synthetic INtron Cassette (SINC), which was then placed downstream of Gmubi and 35S core promoters. Intron fragment tetramers and SINC constructs were evaluated using transient expression in lima bean cotyledons and stable expression in soybean hairy roots. Intron fragments, used as tetramers upstream of the 35S core promoter, yielded up to 80 times higher expression than the core promoter in transient expression analyses and ten times higher expression in stably transformed hairy roots. Tetrameric intronic fragments, cloned downstream of the Gmubi and 35S core promoters and within the synthetic intron, also yielded increased transient and stable GFP expression that was up to 4 times higher than Gmubi alone and up to 40 times higher than the 35S core promoter alone. These intron fragments contain sequences that seem to act as promoter regulatory elements and may contribute to the increased expression observed with this native strong promoter. Intron regulatory elements and synthetic introns may provide additional tools for increasing transgene expression in plants.








Similar content being viewed by others
Abbreviations
- 35S:
-
Cauliflower mosaic virus 35S
- 5′ UTR:
-
5′ Untranslated region
- Gmubi:
-
Glycine max polyubiquitin
- MCS:
-
Multiple cloning site
- MS:
-
Murashige and Skoog
- OMS:
-
MS medium containing no plant growth regulators
- pFLEV:
-
Finer laboratory expression vector
- SINC:
-
Synthetic intron cassette
- TIF:
-
Tetrameric intron fragment
References
Cazzonelli C, Velten J (2007) In vivo characterization of plant promoter element interaction using synthetic promoters. Trans Res 17:437–457
Chen H, Nelson RS, Sherwood JL (1994) Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16:664–670
Chiera JM, Bouchard RA, Dorsey SL, Park E, Buenrostro-Nava MT, Ling PP, Finer JJ (2007) Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26:1501–1509
Chiu WL, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as vital reporter in plants. Curr Biol 6:325–330
Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5´ UTR introns on gene expression in Arabidopsis thaliana. BMC Genom 7:120
De La Torre CM, Finer JJ (2015) The intron and 5′ distal region of the soybean Gmubi promoter contribute to very high levels of gene expression in transiently and stably transformed tissues. Plant Cell Rep 34:111–120
Dean C, Schmidt R (1995) Plant genomes: a current description. Ann Rev Plant Physiol Plant Mol Biol 46:395–418
Finer JJ, Vain P, Jones MW, McMullen MD (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11:323–328
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217:109–119
Hernandez-Garcia CM, Finer JJ (2016) A novel cis-acting element in the GmERF3 promoter contributes to inducible gene expression in soybean and tobacco after wounding. Plant Cell Rep 35:303–316
Hernandez-Garcia CM, Martinelli AP, Bouchard RA, Finer JJ (2009) A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep 28:837–849
Hernandez-Garcia CM, Bouchard RA, Rushton PJ, Jones ML, Chen X, Timko MP, Finer JJ (2010a) High level of transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biol 10:237
Hernandez-Garcia CM, Chiera JM, Finer JJ (2010b) Robotics and dynamic image analysis for studies of gene expression in plant tissues. J Vis Exp. doi:10.3791/1733
Ibraheem O, Botha CEJ, Bradley G (2010) In silico analysis of cis-acting regulatory elements in 5′ regulatory regions of the sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comp Biol Chem 34:268–283
Jeong YM, Mun JH, Kim H, Lee S, Kim SG (2007) An upstream region in the first intron of petunia actin-depolymerizing factor 1 affects tissue-specific expression in transgenic Arabidopsis (Arabidopsis thaliana). Plant J 50:230–239
Kamo K, Kim A, Park SH, Joung YH (2012) The 5′ UTR-intron of the Gladiolus polyubiquitin promoter GUBQ1 enhances translation efficiency in Gladiolus and Arabidopsis. BMC Plant Biol 12:79
Kim MJ, Kim H, Shin JS, Chung C, Ohlrogge JB, Suh MC (2006) Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol Gen Genom 276:351–368
Le Hir H, Nott A, Moore MJ (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28:215–220
Liu W, Mazarei M, Rudis MR, Fethe MH, Stewart CN (2011) Rapid in vivo analysis of synthetic promoters for plant pathogen phytosensing. BMC Biotechnol 11:108
Lu J, Sivamani E, Azhakanandam K, Sammadder P, Xianggan L, Qu R (2008) Gene expression enhancement mediated by the 5′ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Genet Genom 279:563–572
Maniatis T, Tasic B (2002) Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418:236–243
Mehrotra R, Mehrotra S (2010) Promoter activation by ACGT in response to salicylic and abscisic acids is differentially regulated by the spacing between two copies of the motif. J Plant Phys 167:1214–1218
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39:5328–5337
Plesse B, Criqui MC, Durr A, Parmentier Y, Fleck J, Genschik P (2001) Effects of the polyubiquitin gene Ubi.U4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol Biol 45:655–667
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cell Dev Biol Plant 40:1–22
Rasband W (1997) ImageJ. U.S. National Institutes of Health, Bethesda. http://imagej.nih.gov/ij/
Rose AB (2002) Requirements for intron-mediated enhancement of gene expression in Arabidopsis. RNA 8:1444–1453
Rose AB (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40:744–751
Rose AB (2008) Intron-mediated regulation of gene expression. In: Reddy ASN, Golovkin M (eds) Nuclear pre-mRNA processing in plants. Current topics in microbiology and immunology, vol 326. Springer, New York, pp 277–290
Rose AB, Last RL (1997) Introns act post-transcriptionally to increase expression of the Arabidopsis thaliana tryptophan pathway gene PAT1. Plant J 11:455–464
Rose AB, Elfersi T, Parra G, Korf I (2008) Promoter-proximal introns in Arabidopsis are enriched in dispersed signals that elevate gene expression. Plant Cell 20:543–551
Rose AB, Emani S, Bradnam K, Korf I (2011) Evidence for a DNA-based mechanism of intron-mediated enhancement. Frontiers Plant Sci 2:1–9
Rushton PJ, Reinstadler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen-and wound-induced signaling. Plant Cell 14:749–762
Salinas J, Oeda K, Chua N-H (1992) Two G-box-related sequences confer different expression patterns in transgenic tobacco. Plant Cell 4:1485–1493
Samadder P, Sivamani E, Lu J, Li X, Qu R (2008) Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Gen Genom 279:429–439
Sawant SV, Kiran K, Mehrotra R, Chaturvedi CP, Ansari SA, Singh P, Lodhi N, Tuli R (2005) A variety of synergistic and antagonistic interactions mediated by cis-acting DNA motifs regulate gene expression in plant cells and modulate stability of the transcription complex formed on a basal promoter. J Exp Bot 56:2345–2353
Sivamani E, Qu R (2006) Expression enhancement of a rice polyubiquitin gene promoter. Plant Mol Biol 60:225–239
Wang J, Oard JH (2003) Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep 22:129–143
Yang Z, Patra B, Li R, Pattanaik S, Yuan L (2013) Promoter analysis reveals cis-regulatory motifs associated with the expression of the WRKY transcription factor CrWRKY1 in Catharanthus roseus. Planta 238:1039–1049
Zhang N, McHale LK, Finer JJ (2015) Isolation and characterization of “GmScream” promoters that regulate highly expressing soybean (Glycine max Merr.) genes. Plant Sci 241:189–198
Acknowledgements
Salaries and research support were provided by the United Soybean Board, Bayer CropScience, and by State and Federal funds appropriated to the Ohio State University/Ohio Agricultural Research and Development Center. This work was partly supported by an OARDC Director’s Associateship Award to NZ in 2011–2012. This research was also supported, in part, through The Consortium for Plant Biotechnology Research, Inc. by DOE Prime Agreement No. DEFG36-02G012026. This support does not constitute an endorsement by DOE or by the Consortium for Plant Biotechnology Research, Inc. of the views expressed in this publication. Mention of trademark or proprietary products does not constitute a guarantee or warranty of the product by OSU/OARDC and also does not imply approval to the exclusion of other products that may also be suitable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Grant, T.N.L., De La Torre, C.M., Zhang, N. et al. Synthetic introns help identify sequences in the 5′ UTR intron of the Glycine max polyubiquitin (Gmubi) promoter that give increased promoter activity. Planta 245, 849–860 (2017). https://doi.org/10.1007/s00425-016-2646-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-016-2646-8