Abstract
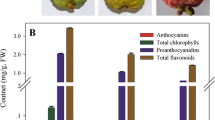
The phenotype of tomato high pigment-1 (hp1) mutant is characterized by overproduction of pigments including chlorophyll and carotenoids during fruit development and ripening. Although the increased plastid compartment size has been thought to largely attribute to the enhanced pigmentation, the molecular aspects of how the HP1/DDB1 gene manipulates plastid biogenesis and development are largely unknown. In the present study, we compared transcriptome profiles of immature fruit pericarp tissue between tomato cv. Ailsa Craig (WT) and its isogenic hp1 mutant. Over 20 million sequence reads, representing > 1.6 Gb sequence data per sample, were generated and assembled into 21,972 and 22,167 gene models in WT and hp1, respectively, accounting for over 60 % official gene models in both samples. Subsequent analyses revealed that 8,322 and 7,989 alternative splicing events, 8833 or 8510 extended 5′-UTRs, 8,263 or 8,939 extended 3′-UTRs, and 1,136 and 1,133 novel transcripts, exist in WT and hp1, respectively. Significant differences in expression level of 880 genes were detected between the WT and hp1, many of which are involved in signaling transduction, transcription regulation and biotic and abiotic stresses response. Distinctly, RNA-seq datasets, quantitative RT-PCR analyses demonstrate that, in hp1 mutant pericarp tissue at early developmental stage, an apparent expression alteration was found in several regulators directly involved in plastid division and development. These results provide a useful reference for a more accurate and more detailed characterization of the molecular process in the development and pigmentation of tomato fruits.







Similar content being viewed by others
Abbreviations
- A5SS:
-
Alternative 5′ splice sites
- A3SS:
-
Alternative 3′ splice sites
- ABA:
-
Abscisic acid
- AFE:
-
Alternative first exons
- ALA:
-
Aminolevulinate
- ALE:
-
Alternative last exons
- DAP:
-
Days after pollination
- DDB1 :
-
UV-DAMAGED DNA BINDING PROTEIN-1
- DET1:
-
DE-ETIOLATED-1
- FDR:
-
False discovery rate
- FPKM:
-
Fragments Per Kilobase of exon per million fragments mapped reads
- GLK:
-
Golden 2-like
- GO:
-
Gene Ontology
- HP :
-
High-Pigment
- MXE:
-
Mutually exclusive exons
- nr databases:
-
Non-redundant databases
- RI:
-
Retained intron
- RIN:
-
RNA Integrity Number
- SE:
-
Skipped exons
- SGN:
-
SOL Genomics Network
- SNP:
-
Single nucleotide polymorphism
- SRA:
-
Sequence read archive
- TSS:
-
Transcription start site
- U :
-
Uniform ripening
- UTRs:
-
Untranslated regions
- WT:
-
Wild type
References
Åkerlund T, Gullbrand B, Nordstrom K (2002) Effects of the Min system on nucleoid segregation in Escherichia coli. Microbiology 148:3213–3222
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Berthe T, Klein-Eude D, Balange AP (2003) Study of 5-aminolevulinate dehydratase in radish seedlings: are there housekeeping and light-induced enzymes? Plant Sci 164:395–405
Bino RJ, Ric de Vos CH, Lieberman M, Hall RD, Bovy A, Jonker HH, Tikunov Y, Lommen A, Moco S, Levin I (2005) The light-hyperresponsive high pigment-2 dg mutation of tomato: alterations in the fruit metabolome. New Phytol 166:427–438
Chi Y, Yang Y, Zhou Y, Zhou J, Fan B, Yu J, Chen Z (2013) Protein-protein interactions in the regulation of WRKY transcription factors. Mol Plant 6:287–300
Chung BY, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5′ UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7:120
Clark TA, Sugner CW, Ares MJ (2002) Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296:907–910
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Cookson PJ, Kiano JW, Shipton CA, Fraser PD, Romer S, Schuch W, Bramley PM, Pyke KA (2003) Increases in cell elongation, plastid compartment size and phytoene synthase activity underlie the phenotype of the high pigment-1 mutant of tomato. Planta 217:896–903
Davuluri GR, Tuinen A, Mustilli AC, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, Pennings HMJ, Bowler C (2004) Manipulation of DET1 expression in tomato results in photomorphogenic phenotypes caused by post-transcriptional gene silencing. Plant J 40:344–354
Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, Bramley PM, Pennings HM, Bowler C (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895
Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J (2008) Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J 53:717–730
Gao H, Kadirjan-Kalbach D, Froehlich JE, Osteryong KW (2003) ARC5, a cytosolic dynamin-like protein from plants, is part of the chloroplast division machinery. Proc Natl Acad Sci USA 100:4328–4333
Gao QM, Venugopal S, Navarre D, Kachroo A (2011) Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol 155:464–476
Giovannoni J (2004) Genetic regulation of fruit development and ripening. Plant Cell 16:S170–S180
Glynn JM, Froehlich JE, Osteryoung KW (2008) Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20:2460–2470
Huang S, Gao Y, Liu J, Peng X, Niu X, Fei Z, Cao S, Liu Y (2012) Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol Genet Genomics 287:495–513
Jarret RL, Sayama H, Tigchelaar EC (1984) Pleiotropic effects associated with the chlorophyll intensifier mutations high pigment and dark green in tomato. J Am Soc Hortic Sci 109:873–878
Jiang H, Wong WH (2009) Statistical inferences for isoform expression in RNA-Seq. Bioinformatics 25:1026–1032
Jones B, Frasse P, Olmos E, Zegzouti H, Li ZG, Latché A, Pech JC, Bouzayen M (2002) Down-regulation of DR12, an auxin-response-factor homolog, in the tomato results in a pleiotropic phenotype including dark green and blotchy ripening fruit. Plant J 32:603–613
Katz Y, Wang ET, Airoldi EM, Burge CB (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7:1009–1015
Kendrick RE, Kerckhoffs LHJ, van Tuinen A, Koornneef M (1997) Photomorphogenic mutants of tomato. Plant, Cell Environ 20:746–751
Kevany BM, Tieman DM, Taylor MG, Dal Cin V, Klee HJ (2008) Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J 51:458–467
Key TJ, Schatzkin A, Willett WC, Allen NE, Spencer EA, Travis RC (2002) Diet, nutrition and the prevention of cancer. Public Health Nutrition 7:187–200
Kim E, Magen A, Ast G (2007) Different levels of alternative splicing among eukaryotes. Nucleic Acids Res 35:125–131
Kolotilin I, Koltai H, Tadmor Y, Bar-Or C, Reuveni M, Meir A, Nahon S, Shlomo H, Chen L, Levin I (2007) Transcriptional profiling of high pigment-2 dg tomato mutant links early fruit plastid biogenesis with its overproduction of phytonutrients. Plant Physiol 145:381–401
Levin I, Frankel P, Gilboa N, Tanny S, Lalazar A (2003) The tomato dark green mutation is a novel allele of the tomato homolog of the DEETIOLATED1 gene. Theor Appl Genet 106:454–460
Lieberman M, Segev O, Gilboa N, Lalazar A, Levin I (2004) The tomato homolog of the gene encoding UV-damaged DNA binding protein 1 (DDB1) underlined as the gene that causes the high pigment-1 mutant phenotype. Theor Appl Genet 108:1574–1581
Liu Y, Roof S, Ye Z, Barry C, van Tuinen A, Vrebalov J, Bowler C, Giovannoni J (2004) Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc Natl Acad Sci USA 101:9897–9902
Liu J, Li H, Miao M, Tang X, Giovannoni J, Xiao F, Liu Y (2012) The tomato UV-damaged DNA-binding protein-1 (DDB1) is implicated in pathogenesis-related (PR) gene expression and resistance to Agrobacterium tumefaciens. Mol Plant Pathol 13:123–134
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P (2008) Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320:789–792
Maple J, Møller SG (2007) Plastid division: evolution, mechanism and complexity. Ann Bot 99:565–579
Maple J, Vojta L, Soll J, Møller SG (2007) ARC3 is a stromal Z-ring accessory protein essential for plastid division. EMBO Rep 8:293–299
Margolin W (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6:862–871
McGuire AM, Pearson MD, Neafsey DE, Galagan JE (2008) Cross-kingdom patterns of alternative splicing and splice recognition. Genome Biol 9:R50
Mignone F, Gissi C, Liuni S, Pesole G (2002) Untranslated regions of mRNAs. Genome Biol 3: reviews0004.1-0004.10
Miyagishima S, Nishida K, Mori T, Matsuzaki M, Higashiyama T, Kuroiwa H, Kuroiwa T (2003) A plant-specific dynamin-related protein forms a ring at the chloroplast division site. Plant Cell 15:655–665
Miyagishima SY, Froehlich JE, Osteryoung KW (2006) PDV1 and PDV2 mediate recruitment of the dynamin-related protein ARC5 to the plastid division site. Plant Cell 18:2517–2530
Modrek B, Lee C (2002) A genomic view of alternative splicing. Nat Genet 30:13–19
Mori T, Kuroiwa H, Takahara M, Miyagishima SY, Kuroiwa T (2001) Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol 42:555–559
Mustilli AC, Fenzi F, Ciliento R, Alfano F, Bowler C (1999) Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11:145–157
Nagata N, Tanaka R, Satoh S, Tanaka A (2005) Identification of a vinyl reductase gene for chlorophyll synthesis in Arabidopsis thaliana and implications for the evolution of Prochlorococcus species. Plant Cell 17:223–240
Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39:877–885
Nicolae M, Mangul S, Măndoiu II, Zelikovsky A (2011) Estimation of alternative splicing isoform frequencies from RNA-Seq data. Algorithm Mol Biol 6:9
Osawa M, Anderson DE, Erickson HP (2008) Reconstitution of contractile FtsZ rings in liposomes. Science 320:792–794
Osteryoung KW, Vierling E (1995) Conserved cell and organelle division. Nature 376:473–474
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Papenbrock J, Pfündel E, Mock HP, Grimm B (2000) Decreased and increased expression of the subunit CHL I diminishes Mg chelatase activity and reduces chlorophyll synthesis in transgenic tobacco plants. Plant J 22:155–164
Pepper A, Delaney T, Washburn T, Poole D, Chory J (1994) DET1, a negative regulator of light-mediated development and gene expression in arabidopsis, encodes a novel nuclear-localized protein. Cell 78:109–116
Powell AL, Nguyen CV, Hill T, Cheng KL, Figueroa-Balderas R, Aktas H, Ashrafi H, Pons C, Fernández-Muñoz R, Vicente A, Lopez-Baltazar J, Barry CS, Liu YS, Chetelat R, Granell A, Van Deynze A, Giovannoni JJ, Bennett AB (2012) Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 336:1711–1715
Raynaud C, Perennes C, Reuzeau C, Catrice O, Brown S, Bergounioux C (2005) Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc Natl Acad Sci USA 102:8216–8221
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Schmitz AJ, Glynn JM, Olson BJ, Stokes KD, Osteryoung KW (2009) Arabidopsis FtsZ2-1 and FtsZ2-2 are functionally redundant, but FtsZ-based plastid division is not essential for chloroplast partitioning or plant growth and development. Mol Plant 2:1211–1222
Schroeder DF, Gahrtz M, Maxwell BB, Cook RK, Kan JM, Alonso JM, Ecker JR, Chory J (2002) De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr Biol 12:1462–1472
Shalygo N, Czarnecki O, Peter E, Grimm B (2009) Expression of chlorophyll synthase is also involved in feedback-control of chlorophyll biosynthesis. Plant Mol Biol 71:425–436
Shimada H, Koizumi M, Kuroki K, Mochizuki M, Fujimoto H, Ohta H, Masuda T, Takamiya K (2004) ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol 45:960–967
Spiegel-Roy P, Goldschmidt EE (1996) Fruit development and maturation. In: Spiegel-Roy P, Goldschmidt EE (eds) Biology of horticultural crops. Cambridge University Press, Great Britain, pp 92–107
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, van Baren M, Salzberg S, Wold B, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Van den Berg N, Berger DK, Hein I, Birch PR, Wingfield MJ, Viljoen A (2007) Tolerance in banana to Fusarium wilt is associated with early up-regulation of cell wall-strengthening genes in the roots. Mol Plant Pathol 8:333–341
Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153:111–120
Vitha S, Froehlich JE, Koksharova O, Pyke KA, van Erp H, Osteryoung KW (2003) ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 15:1918–1933
Wang S, Liu J, Feng Y, Niu X, Giovannoni J, Liu Y (2008) Altered plastid levels and potential for improved fruit nutrient content by downregulation of the tomato DDB1-interacting protein CUL4. Plant J 55:89–103
Wang H, Schauer N, Usadel B, Frasse P, Zouine M, Hernould M, Latche A, Pech JC, Fernie AR, Bouzayen M (2009) Regulatory features underlying pollination-dependent and -independent tomato fruit set revealed by transcript and primary metabolite profiling. Plant Cell 21:1428–1452
Wang L, Feng Z, Wang X, Wang X, Zhang X (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138
Yen HC, Shelton BA, Howard LR, Lee S, Vrebalov J, Giovannoni JJ (1997) The tomato high-pigment (hp) locus maps to chromosome 2 and influences plastome copy number and fruit quality. Theor Appl Genet 95:1069–1079
Yoder DW, Kadirjan-Kalbach D, Olson BJ, Miyagishima SY, Deblasio SL, Hangarterr RP, Osteryong KW (2007) Effects of mutations in Arabidopsis FtsZ1 on plastid division, FtsZ ring formation and positioning, and FtsZ filament morphology in vivo. Plant Cell Physiol 48:775–791
Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, Malerba G, Bellin D, Pezzotti M, Delledonne M (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152:1787–1795
Acknowledgments
This work was supported by the National Science Fund for Distinguished Young Scholars (No. 30825030), the National Science and Technology Key Project of China (No. 2011CB100401), the National Natural Science Foundation of China (No. 31171179), and Advanced Program of Doctoral Fund of Ministry of Education of China (20110181130009) for Y. Liu, and the Fundamental Research Funds for the Central Universities for S. Huang.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xiaofeng Tang, Zizhi Tang, Shengxiong Huang, and Jikai Liu contributed equally.
Electronic supplementary material
Below is the link to the electronic supplementary material.
425_2013_1942_MOESM1_ESM.doc
Supplemental Table S1 The sequences of specific primers used in RT-PCR for validation of novel untranslated region, novel transcripts, and alternative splicing isoforms. “Locus” shows the exact location of each gene relative to S. lycopersicum genome (the SL2.40 version). “Gene model” displays the corresponding official gene ID in SGN (http://solgenomics.net). Primer A (upper) and primer B (lower) represent the positive primer for detected gene sequence in official gene models and determined by RNA-Seq, respectively. Primer C represents the negative primer sequence (DOC 49 kb)
425_2013_1942_MOESM2_ESM.doc
Supplemental Table S2 Primer sequences used in RT-PCR validation of differentially expressed genes involved in chlorophyll metabolism, plastid division and other biological processes in immature fruits of WT and hp1 mutant. “Gene” and “Gene model” represent the gene symbol and corresponding official gene ID in SGN (http://solgenomics.net), respectively(DOC 69 kb)
425_2013_1942_MOESM3_ESM.xls
Supplemental Table S3 Annotation results of transcriptome assembly. The “Gene model” stands for the corresponding official gene ID in SGN (http://solgenomics.net). “Direction” represents the positive/negative strand of DNA strand. “FPKM of WT” and “FPKM of hp1” means FPKM value of the transcripts in WT and hp1 mutant, respectively. The “log (fold_change)” is the value of log (FPKM of hp1/FPKM of WT). The “p_value” is calculated by the Fisher’s exact test. “Seq_Description”, “Seq_Length”, “GOs”, “Enzyme_Codes” and “sequence” are the annotation for putative gene function, length, GO annotation, enzyme annotation and the exact nucleotide sequence, respectively. “--NA--” indicates the transcripts in WT or hp1 mutant without significant hit(XLS 31165 kb)
425_2013_1942_MOESM4_ESM.xls
Supplemental Table S4 Statistical results of indentified and extended untranslated regions (UTRs). The sheet “WT_5” and “WT_3” displays statistical results of indentified and extended 5′- and 3′- UTRs of transcripts in WT, respectively. The sheet “hp1_5” and “hp1_3” displays statistical results of indentified and extended 5′- and 3′- UTRs of transcripts in hp1 mutant, respectively. In each sheet, the following contents are included. The “Gene model” represents official gene ID in SGN (http://solgenomics.net). “identified UTRs” shows the length of identified 5′ or 3′ end boundaries, and “extended UTRs” shows the length of extended 5′ or 3′ end boundaries which are extended relative to the end of official gene models in WT, respectively(XLS 3390 kb)
425_2013_1942_MOESM5_ESM.xls
Supplemental Table S5 Statistical results of five types of alternative splicing events. “WT only”, “hp1 only” and “WT&hp1” stand for the alternative splicing events in AC, hp1 mutant and both genotypes, respectively. “Gene model” displays the corresponding official gene ID in SGN (http://solgenomics.net). “Alternative splicing events” indicates specific alternative splicing events for each gene(XLS 1712 kb)
425_2013_1942_MOESM6_ESM.xls
Supplemental Table S6 Statistical results of novel transcripts. “WT”, “hp1” and “WT&hp1″ stand for the novel transcripts in WT, hp1 mutant and both genotypes, respectively. “Gene model” displays the corresponding official gene ID in SGN (http://solgenomics.net). “Direction” represents the positive or negative direction of the DNA strand. “Locus” shows the exact location of each gene relative to S. lycopersicum genome (the SL2.40 version). “Seq_Description”, “Seq_Length” and “sequence” represent the annotation for putative gene function, length, and the exact nucleotide sequence, respectively. “--NA--” indicates the transcripts in WT or hp1 mutant without significant hit, comparing to the official annotation (nr database) (XLS 1274 kb)
425_2013_1942_MOESM7_ESM.xls
Supplemental Table S7 Genes differentially expressed in the WT and hp1 mutant. The sheets “up diff” and “down diff” indicate genes were significantly up-regulated or down-regulated in the hp1 mutant compared to those in the WT, respectively (p < 0.05). “Gene model” represents the corresponding official gene ID in SGN (http://solgenomics.net). “Locus” shows the exact location of each gene relative to S. lycopersicum genome (the SL2.40 version). “FPKM of WT” and “FPKM of hp1” stand for the FPKM value of the transcripts in WT and hp1 mutant, respectively. The “ln (fold_change)” is the value of ln (FPKM of hp1/FPKM of WT). The “p_value” is determined by the Student’s t-test(XLS 548 kb)
Rights and permissions
About this article
Cite this article
Tang, X., Tang, Z., Huang, S. et al. Whole transcriptome sequencing reveals genes involved in plastid/chloroplast division and development are regulated by the HP1/DDB1 at an early stage of tomato fruit development. Planta 238, 923–936 (2013). https://doi.org/10.1007/s00425-013-1942-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-013-1942-9