Abstract
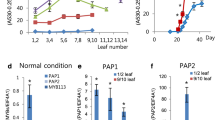
Expression of regulators of the flavonoid pathway was examined in Arabidopsis thaliana wild type and pap1D plants, the latter being a T-DNA activation-tagged line over-expressing the PAP1/MYB75 gene which is a positive regulator of the pathway. Anthocyanin accumulation was induced in plants grown in soil, on agar plates, and hydroponics by withdrawing nitrogen from the growth medium. The agar-grown seedlings and rosette stage plants in hydroponics were further explored, and showed that nitrogen deficiency resulted in the accumulation of not only anthocyanins, but also flavonols. The examination of transcript levels showed that the general flavonoid pathway regulators PAP1 and PAP2 were up-regulated in response to nitrogen deficiency in wild type as well as pap1D plants. Interestingly, PAP2 responded much stronger to nitrogen deficiency than PAP1, 200- and 6-fold increase in transcript levels, respectively, for wild-type seedlings. In rosette leaves the increase was 900-fold for PAP2 and 6-fold for PAP1. At least three different bHLH domain transcription factors promote anthocyanin synthesis, and transcripts for one of these, i.e. GL3 were found to be sixfold enhanced by nitrogen deficiency in rosette leaves. The MYB12 transcription factor, known to regulate flavonol synthesis, was slightly induced by nitrogen deficiency in seedlings. In conclusion, four out of eight regulators involved in the flavonoid pathway showed an enhanced expression from 2 to 1,000 times in response to nitrogen deficiency. Together with MYB factors, especially PAP2, GL3 appears to be the BHLH partner for anthocyanin accumulation in response to nitrogen deficiency.



Similar content being viewed by others
Abbreviations
- BHLH:
-
Basic helix-loop-helix domain
- EGL3:
-
Enhancer of GLABRA3
- GL3:
-
GLABRA3
- MYB:
-
DNA-binding domain
- PAL:
-
Phenylalanine ammonium lyase
- PAP1(2):
-
Production of anthocyanin pigment 1 (2)
- TTG1:
-
Transparent TESTA GLABRA1
- TT8:
-
Transparent TESTA8
- WT:
-
Wild type
References
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12:2383–2393
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Ecp Stn Circular 347:1–32
Hsieh M-H, Lam H-M, van de Loo F, Coruzzi G (1998) A PII-like protein in Arabidopsis: Putative role in nitrogen sensing. Proc Natl Acad Sci USA 95:13965–13970
Kliebenstein DJ (2004) Secondary metabolites and plant/environment interactions: a view through Arabidopsis thaliana tinged glasses. Plant Cell Environ 27:675–684
Koes R, Verweij W, Quattrocchio F (2005) Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci 10:236–242
Krapp A, Truong H-N (2006) Regulation of C/N interaction in model species. In: Goyal SS, Tischner R, Basra AS (eds) Enhancing the efficiency of nitrogen utilization in plants. The Haworth Press, New York, pp 127–173
Lehfeldt CU (2001) Das Gen der Sinapoylglucose:L-Malat-Sinapoyltransferase von Arabidopsis thaliana (L.) Heynh. (Ackerschmalwand): Klonierung durch T-DNA-Tagging und Versuche zur Expression in E.coli. PhD-Dissertation. Martin Luther Universität, Halle-Wittenberg
Lloyd JC, Zakhleniuk OV (2004) Responses of primary and secondary metabolism to sugar accumulation revealed by microarray expression analysis of the Arabidopsis mutant, pho3. J Exp Bot 55:1221–1230
Mabry TJ, Markham KR, Thomas MB (1970) The systematic identification of flavonoids. Springer, Berlin Heidelberg New York
Markham KR (1982) Techniques of flavonoids identification. Academic, London
Martens S, Knott J, Seitz CA, Janvari L, Yu S-N, Forkmann G (2003) Impact of biochemical pre-studies on specific metabolic engineering strategies of flavonoid biosynthesis in plant tissue. Biochem Eng J 14:227–235
Martin T, Oswald O, Graham IA (2002) Arabidopsis seedling growth, storage lipid mobilization, and photosynthetic gene expression are regulated by carbon:nitrogen availability. Plant Physiol 128:472–481
Mehrtens F, Kranz H, Bednarek P, Weisshaar B (2005) The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol 138:1083–1096
Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A (2006) Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224:556–568
Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Doorsselaere JV, Joseleau J-P, Vuylsteke M, Van Driessche G, Van Beeumen J, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16:2749–2771
Scheible W-R, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopis in response to nitrogen. Plant Physiol 136:2483–2499
Schijlen EGWM, Ric deVos CH, van Tunen AJ, Bovy AG (2004) Modification of flavonoid biosynthesis in crop plants. Phytochemistry 65:2631–2648
Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8:659–671
Smith CS, Weljie AM, Moorhead GB (2003) Molecular properties of the putative nitrogen sensor PII from Arabidopis thaliana. Plant J 33:353–360
Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140:637–646
Stewart AJ, Chapman W, Jenkins GI, Graham I, Martin T, Crozier A (2001) The effect of nitrogen and phosphorus deficiency on flavonol accumulation in plant tissues. Plant Cell Environ 24:1189–1197
Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139:1840–1852
Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J-i, Awazuhara M, Inoue E, Tkahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, Saito K (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42:218–235
Veit M, Pauli GF (1999) Major flavonoids from Arabidopsis thaliana leaves. J Nat Prod 62:1301–1303
Wade HK, Sohal AK, Jenkins GI (2003) Arabidopsis ICX1 is a negative regulator of several pathways regulating flavonoid biosynthesis genes. Plant Physiol 131:707–715
Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford N (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopis. Plant Physiol 136:2512–2522
Weaver LM, Herrmann KM (1997) Dynamics of the shikimate pathway in plants. Trends Plant Sci 2:346–351
Wintersohl U, Krause J, Napp-Zinn K (1979) Phenylpropane derivatives in Arabidopsis thaliana (L.) Heynh. Arabidopsis Inf Serv 16:76–77
Zhang F, Gonzalez A, Zhao M, Payne T, Lloyd A (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130:4859–4869
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40:22–34
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lea, U.S., Slimestad, R., Smedvig, P. et al. Nitrogen deficiency enhances expression of specific MYB and bHLH transcription factors and accumulation of end products in the flavonoid pathway. Planta 225, 1245–1253 (2007). https://doi.org/10.1007/s00425-006-0414-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-006-0414-x