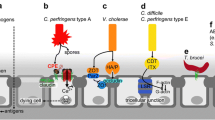
Abstract
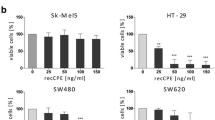
Given that most malignant tumors are derived from epithelium, developing a strategy for treatment of epithelium-derived cancers (i.e., carcinomas) is a pivotal issue in cancer therapy. Carcinomas, including ovarian, breast, prostate, and pancreatic cancers, are known to overexpress various claudins (CLDNs); in particular, CLDN-3 and -4 are frequently overexpressed in malignant case. The generation of CLDN binders is a key for expanding CLDN-targeted cancer therapy but has been delayed due to the small size of CLDN extracellular domains (approximately 50 amino acids for the first domain and 15 amino acids for the second) and their high homology among species. Interestingly, however, the receptors for Clostridium perfringens enterotoxin (CPE), a foodborne toxin in humans, happen to be identical to CLDN-3 and -4. Thus, the first CLDN binder, CPE, has provided us CLDN-targeted cancer therapy from a concept into a potential reality. In this review, we describe roles of CPE technology in cancer therapy and discuss future directions in the CLDN-targeting concept-to-therapy process.




Similar content being viewed by others
References
Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI, Guseh JS, Rajagopal J, Melton DA (2008) Genetic targeting of the endoderm with Claudin-6(CreER). Dev Dynam 237:504–512
Baronzio G, Parmar G, Baronzio M (2015) Overview of methods for overcoming hindrance to drug delivery to tumors, with special attention to tumor interstitial fluid. Front Oncol 5:165
Ben-David U, Nudel N, Benvenisty N (2013) Immunologic and chemical targeting of the tight-junction protein claudin-6 eliminates tumorigenic human pluripotent stem cells. Nat Commun 4
Beyer I, Cao H, Persson J, Song H, Richter M, Feng Q, Yumul R, van Rensburg R, Li Z, Berenson R, Carter D, Roffler S, Drescher C, Lieber A (2012) Coadministration of epithelial junction opener JO-1 improves the efficacy and safety of chemotherapeutic drugs. Clin Cancer Res 18:3340–3351
Beyer I, van Rensburg R, Strauss R, Li Z, Wang H, Persson J, Yumul R, Feng Q, Song H, Bartek J, Fender P, Lieber A (2011) Epithelial junction opener JO-1 improves monoclonal antibody therapy of cancer. Cancer Res 71:7080–7090
Blanchard AA, Skliris GP, Watson PH, Murphy LC, Penner C, Tomes L, Young TL, Leygue E, Myal Y (2009) Claudins 1, 3, and 4 protein expression in ER negative breast cancer correlates with markers of the basal phenotype. Virchows Arch 454:647–656
Borlak J, Meier T, Halter R, Spanel R, Spanel-Borowski K (2005) Epidermal growth factor-induced hepatocellular carcinoma: gene expression profiles in precursor lesions, early stage and solitary tumours. Oncogene 24:1809–1819
Casagrande F, Cocco E, Bellone S, Richter CE, Bellone M, Todeschini P, Siegel E, Varughese J, Arin-Silasi D, Azodi M, Rutherford TJ, Pecorelli S, Schwartz PE, Santin AD (2011) Eradication of chemotherapy-resistant CD44+ human ovarian cancer stem cells in mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer-. Am Cancer Soc 117:5519–5528
Chakrabarti G, McClane BA (2005) The importance of calcium influx, calpain and calmodulin for the activation of CaCo-2 cell death pathways by Clostridium perfringens enterotoxin. Cell Microbiol 7:129–146
Chen J, Theoret JR, Shrestha A, Smedley JG 3rd, McClane BA (2012) Cysteine-scanning mutagenesis supports the importance of Clostridium perfringens enterotoxin amino acids 80 to 106 for membrane insertion and pore formation. Infect Immun 80:4078–4088
Cheung ST, Leung KL, Ip YC, Chen X, Fong DY, Ng IO, Fan ST, So S (2005) Claudin-10 expression level is associated with recurrence of primary hepatocellular carcinoma. Clin Cancer Res 11:551–556
Cocco E, Shapiro EM, Gasparrini S, Lopez S, Schwab CL, Bellone S, Bortolomai I, Sumi NJ, Bonazzoli E, Nicoletti R, Deng Y, Saltzman WM, Zeiss CJ, Centritto F, Black JD, Silasi DA, Ratner E, Azodi M, Rutherford TJ, Schwartz PE, Pecorelli S, Santin AD (2015) Clostridium perfringens enterotoxin C-terminal domain labeled to fluorescent dyes for in vivo visualization of micrometastatic chemotherapy-resistant ovarian cancer. Int J Cancer 137:2618–2629
Davidson B, Zhang Z, Kleinberg L, Li M, Florenes VA, Wang TL, Shih IM (2006) Gene expression signatures differentiate ovarian/peritoneal serous carcinoma from diffuse malignant peritoneal mesothelioma. Clin Cancer Res 12:5944–5950
de Oliveira SS, de Oliveira IM, De Souza W, Morgado-Diaz JA (2005) Claudins upregulation in human colorectal cancer. FEBS Lett 579:6179–6185
DeFeo-Jones D, Garsky VM, Wong BK, Feng DM, Bolyar T, Haskell K, Kiefer DM, Leander K, McAvoy E, Lumma P, Wai J, Senderak ET, Motzel SL, Keenan K, Van Zwieten M, Lin JH, Freidinger R, Huff J, Oliff A, Jones RE (2000) A peptide-doxorubicin prodrug activated by prostate-specific antigen selectively kills prostate tumor cells positive for prostate-specific antigen in vivo. Nat Med 6:1248–1252
dos Reis PP, Bharadwaj RR, Machado J, MacMillan C, Pintilie M, Sukhai MA, Perez-Ordonez B, Gullane P, Irish J, Kamel-Reid S (2008) Claudin 1 overexpression increases invasion and is associated with aggressive histological features in oral squamous cell carcinoma. Cancer-Am Cancer Soc 113:3169–3180
Fujita K, Katahira J, Horiguchi Y, Sonoda N, Furuse M, Tsukita S (2000) Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett 476:258–261
Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S (1998) Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol 141:1539–1550
Furuse M, Sasaki H, Fujimoto K, Tsukita S (1998) A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J Cell Biol 143:391–401
Gao Z, Xu X, McClane B, Zeng Q, Litkouhi B, Welch WR, Berkowitz RS, Mok SC, Garner EI (2011) C terminus of Clostridium perfringens enterotoxin downregulates CLDN4 and sensitizes ovarian cancer cells to taxol and carboplatin. Clin Cancer Res 17:1065–1074
Grass JE, Gould LH, Mahon BE (2013) Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998–2010. Foodborne Pathog Dis 10:131–136
Grone J, Weber B, Staub E, Heinze M, Klaman I, Pilarsky C, Hermann K, Castanos-Velez E, Ropcke S, Mann B, Rosenthal A, Buhr HJ (2007) Differential expression of genes encoding tight junction proteins in colorectal cancer: frequent dysregulation of claudin-1, -8 and -12. Int J Color Dis 22:651–659
Gunzel D, Fromm M (2012) Claudins and other tight junction proteins. Compr Physiol 2:1819–1852
Hanna PC, Mietzner TA, Schoolnik GK, Mcclane BA (1991) Localization of the receptor-binding region of clostridium-perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides—the 30 C-terminal amino-acids define a functional binding region. J Biol Chem 266:11037–11043
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu ZY, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin YZ, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, and Perou CM (2007) Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8:
Higashi Y, Suzuki S, Sakaguchi T, Nakamura T, Baba S, Reinecker HC, Nakamura S, Konno H (2007) Loss of claudin-1 expression correlates with malignancy of hepatocellular carcinoma. J Surg Res 139:68–76
Hofmann HS, Bartling B, Simm A, Murray R, Aziz N, Hansen G, Silber RE, Burdach S (2006) Identification and classification of differentially expressed genes in non-small cell lung cancer by expression profiling on a global human 59.620-element oligonucleotide array. Oncol Rep 16:587–595
Hornsby CD, Cohen C, Amin MB, Picken MM, Lawson D, Yin-Goen QQ, Young AN (2007) Claudin-7 immunohistochemistry in renal tumors—a candidate marker for chromophobe renal cell carcinoma identified by gene expression profiling. Arch Pathol Lab Med 131:1541–1546
Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ (2000) Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res 60:6281–6287
Karanjawala ZE, Illei PB, Ashfaq R, Infante JR, Murphy K, Pandey A, Schulick R, Winter J, Sharma R, Maitra A, Goggins M, Hruban RH (2008) New markers of pancreatic cancer identified through differential gene expression analyses: claudin 18 and annexin A8. Am J Surg Pathol 32:188–196
Katahira J, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N (1997) Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J Cell Biol 136:1239–1247
Katahira J, Sugiyama H, Inoue N, Horiguchi Y, Matsuda M, Sugimoto N (1997) Clostridium perfringens enterotoxin utilizes two structurally related membrane proteins as functional receptors in vivo. J Biol Chem 272:26652–26658
Kim TH, Huh JH, Lee S, Kang H, Kim GI, An HJ (2008) Down-regulation of claudin-2 in breast carcinomas is associated with advanced disease. Histopathology 53:48–55
Kinugasa T, Huo Q, Higash D, Shibaguchi H, Kuroki M, Tanaka T, Futami K, Yamashita Y, Hachimine K, Maekawa S, Nabeshima K, Iwasaki H, Kuroki M (2007) Selective up-regulation of claudin-1 and claudin-2 in colorectal cancer. Anticancer Res 27:3729–3734
Kitadokoro K, Nishimura K, Kamitani S, Fukui-Miyazaki A, Toshima H, Abe H, Kamata Y, Sugita-Konishi Y, Yamamoto S, Karatani H, Horiguchi Y (2011) Crystal structure of Clostridium perfringens enterotoxin displays features of beta-pore-forming toxins. J Biol Chem 286:19549–19555
Kokai-Kun JF, Benton K, Wieckowski EU, McClane BA (1999) Identification of a Clostridium perfringens enterotoxin region required for large complex formation and cytotoxicity by random mutagenesis. Infect Immun 67:5634–5641
Kominsky SL (2006) Claudins: emerging targets for cancer therapy. Expert Rev Mol Med 8:1–11
Kominsky SL, Vali M, Korz D, Gabig TG, Weitzman SA, Argani P, Sukumar S (2004) Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am J Pathol 164:1627–1633
Kondoh M, Masuyama A, Takahashi A, Asano N, Mizuguchi H, Koizumi N, Fujii M, Hayakawa T, Horiguchi Y, Watanbe Y (2005) A novel strategy for the enhancement of drug absorption using a claudin modulator. Mol Pharmacol 67:749–756
Konecny GE, Agarwal R, Keeney GA, Winterhoff B, Jones MB, Mariani A, Riehle D, Neuper C, Dowdy SC, Wang HJ, Morin PJ, Podratz KC (2008) Claudin-3 and claudin-4 expression in serous papillary, clear-cell, and endometrioid endometrial cancer. Gynecol Oncol 109:263–269
Kuhn S, Koch M, Nubel T, Ladwein M, Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L, Franke WW, Weitz J, Zoller M (2007) A complex of EpCAM, claudin-7, CD44 variant isoforms, and tetraspanins promotes colorectal cancer progression. Mol Cancer Res 5:553–567
Kyuno D, Yamaguchi H, Ito T, Kono T, Kimura Y, Imamura M, Konno T, Hirata K, Sawada N, Kojima T (2014) Targeting tight junctions during epithelial to mesenchymal transition in human pancreatic cancer. World J Gastroentero 20:10813–10824
Landers KA, Samaratunga H, Teng L, Buck M, Burger MJ, Scells B, Lavin MF, Gardiner RA (2008) Identification of claudin-4 as a marker highly overexpressed in both primary and metastatic prostate cancer. Brit. J Cancer 99:491–501
Leach MW, Rottman JB, Hock MB, Finco D, Rojko JL, Beyer JC (2014) Immunogenicity/hypersensitivity of biologics. Toxicol Pathol 42:293–300
Lechpammer M, Resnick MB, Sabo E, Yakirevich E, Greaves WO, Sciandra KT, Tavares R, Noble LC, DeLellis RA, Wang LJ (2008) The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Modern Pathol 21:1320–1329
Lee AS, Tang C, Rao MS, Weissman IL, Wu JC (2013) Tumorigenicity as a clinical hurdle for pluripotent stem cell therapies. Nat Med 19:998–1004
Lee JW, Lee SJ, Seo J, Song SY, Ahn G, Park CS, Lee JH, Kim BG, Bae DS (2005) Increased expressions of claudin-1 and claudin-7 during the progression of cervical neoplasia. Gynecol Oncol 97:53–59
Leech AO, Cruz RG, Hill AD, Hopkins AM (2015) Paradigms lost-an emerging role for over-expression of tight junction adhesion proteins in cancer pathogenesis. Ann Transl Med 3:184
Li XR, Saeki R, Watari A, Yagi K, Kondoh M (2014) Tissue distribution and safety evaluation of a claudin-targeting molecule, the C-terminal fragment of Clostridium perfringens enterotoxin. Eur J Pharm Sci 52:132–137
Ling J, Liao H, Clark R, Wong MS, Lo DD (2008) Structural constraints for the binding of short peptides to claudin-4 revealed by surface plasmon resonance. J Biol Chem 283:30585–30595
Long HY, Crean CD, Lee WH, Cummings OW, Gabig TG (2001) Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res 61:7878–7881
Maeda T, Murata M, Chiba H, Takasawa A, Tanaka S, Kojima T, Masumori N, Tsukamoto T, Sawada N (2012) Claudin-4-targeted therapy using Clostridium perfringens enterotoxin for prostate cancer. Prostate 72:351–360
Marcucci F, Corti A (2012) Improving drug penetration to curb tumor drug resistance. Drug Discov Today 17:1139–1146
Michl P, Buchholz M, Rolke M, Kunsch S, Lohr M, McClane B, Tsukita S, Leder G, Adler G, Gress TM (2001) Claudin-4: a new target for pancreatic cancer treatment using Clostridium perfringens enterotoxin. Gastroenterology 121:678–684
Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, Tsukita S (2011) Predicted expansion of the claudin multigene family. FEBS Lett 585:606–612
Moldvay J, Jackel M, Paska C, Soltesz I, Schaff Z, Kiss A (2007) Distinct claudin expression profile in histologic subtypes of lung cancer. Lung Cancer 57:159–167
Montgomery E, Mamelak AJ, Gibson M, Maitra A, Sheikh S, Amr SS, Yang S, Brock M, Forastiere A, Zhang S, Murphy KM, Berg KD (2006) Overexpression of claudin proteins in esophageal adenocarcinoma and its precursor lesions. Appl Immunohisto M M 14:24–30
Morita K, Furuse M, Fujimoto K, Tsukita S (1999) Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. P Natl Acad Sci USA 96:511–516
Mosley M, Knight J, Neesse A, Michl P, Iezzi M, Kersemans V, Cornelissen B (2015) Claudin-4 SPECT imaging allows detection of aplastic lesions in a mouse model of breast cancer. J Nucl Med 56:745–751
Nacht M, Ferguson AT, Zhang W, Petroziello JM, Cook BP, Gao YH, Maguire S, Riley D, Coppola G, Landes GM, Madden SL, Sukumar S (1999) Combining serial analysis of gene expression and array technologies to identify genes differentially expressed in breast cancer. Cancer Res 59:5464–5470
Nakanishi K, Ogata S, Hiroi S, Tominaga S, Aida S, Kawai T (2008) Expression of occludin and claudins 1, 3, 4, and 7 in urothelial carcinoma of the upper urinary tract. Am J Clin Pathol 130:43–49
Neesse A, Hahnenkamp A, Griesmann H, Buchholz M, Hahn SA, Maghnouj A, Fendrich V, Ring J, Sipos B, Tuveson DA, Bremer C, Gress TM, Michl P (2013) Claudin-4-targeted optical imaging detects pancreatic cancer and its precursor lesions. Gut 62:1034–1043
Nichols LS, Ashfaq R, Iacobuzio-Donahue CA (2004) Claudin 4 protein expression in primary and metastatic pancreatic cancer—support for use as a therapeutic target. Am J Clin Pathol 121:226–230
Oliveira SS, Morgado-Diaz JA (2007) Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci 64:17–28
Park JY, Park KH, TY O, Hong SP, Jeon TJ, Kim CH, Park SW, Chung JB, Song SY, Bang S (2007) Up-regulated claudin 7 expression in intestinal-type gastric carcinoma. Oncol Rep 18:377–382
Protze J, Eichner M, Piontek A, Dinter S, Rossa J, Blecharz KGZ, Vajkoczy P, Piontek J, Krause G (2015) Directed structural modification of Clostridium perfringens enterotoxin to enhance binding to claudin-5. Cell Mol Life Sci 72:1417–1432
Resnick MB, Gavilanez M, Newton E, Konkin T, Bhattacharya B, Britt DE, Sabo E, Moss SF (2005) Claudin expression in gastric adenocarcinomas: a tissue microarray study with prognostic correlation. Hum Pathol 36:886–892
Resnick MB, Konkin T, Routhier J, Sabo E, Pricolo VE (2005) Claudin-1 is a strong prognostic indicator in stage II colonic cancer: a tissue microarray study. Modern Pathol 18:511–518
Romanov V, Whyard TC, Waltzer WC, Gabig TG (2014) A claudin 3 and claudin 4-targeted Clostridium perfringens protoxin is selectively cytotoxic to PSA-producing prostate cancer cells. Cancer Lett 351:260–264
Sahin U, Koslowski M, Dhaene K, Usener D, Brandenburg G, Seitz G, Huber C, Tureci O (2008) Claudin-18 splice variant 2 is a pan-cancer target suitable for therapeutic antibody development. Clin Cancer Res 14:7624–7634
Saitoh Y, Suzuki H, Tani K, Nishikawa K, Irie K, Ogura Y, Tamura A, Tsukita S, Fujiyoshi Y (2015) Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347:775–778
Sanada Y, Oue N, Mitani Y, Yoshida K, Nakayama H, Yasui W (2006) Down-regulation of the claudin-18 gene, identified through serial analysis of gene expression data analysis, in gastric cancer with an intestinal phenotype. J Pathol 208:633–642
Santin AD, Bellone S, Siegel ER, McKenney JK, Thomas M, Roman JJ, Burnett A, Tognon G, Bandiera E, Pecorelli S (2007) Overexpression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in uterine carcinosarcomas. Clin Cancer Res 13:3339–3346
Santin AD, Cane S, Bellone S, Palmieri M, Siegel ER, Thomas M, Roman JJ, Burnett A, Cannon MJ, Pecorelli S (2005) Treatment of chemotherapy-resistant human ovarian cancer xenografts in C.B-17/SCID mice by intraperitoneal administration of Clostridium perfringens enterotoxin. Cancer Res 65:4334–4342
Santin AD, Zhan FH, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O’Brien TJ, Tian EM, Cannon MJ, Shaughnessy J, Pecorelli S (2004) Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer 112:14–25
Shang X, Lin X, Manorek G, Howell SB (2013) Claudin-3 and claudin-4 regulate sensitivity to cisplatin by controlling expression of the copper and cisplatin influx transporter CTR1. Mol Pharmacol 83:85–94
Sobel G, Paska C, Szabo I, Kiss A, Kadar A, Schaff Z (2005) Increased expression of claudins in cervical squamous intraepithelial neoplasia and invasive carcinoma. Hum Pathol 36:162–169
Soini Y, Talvensaari-Mattila A (2006) Expression of claudins 1, 4, 5, and 7 in ovarian tumors of diverse types. Int J Gynecol Pathol 25:330–335
Sonoda N, Furuse M, Sasaki H, Yonemura S, Katahira J, Horiguchi Y, Tsukita S (1999) Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: evidence for direct involvement of claudins in tight junction barrier. J Cell Biol 147:195–204
Sugii S (1994) Analysis of multiple antigenic determinants of clostridium-perfringens enterotoxin as revealed by use of different synthetic peptides. J Vet Med Sci 56:1047–1050
Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y (2014) Crystal structure of a claudin provides insight into the architecture of tight junctions. Science 344:304–307
Szasz AM, Nyirady P, Majoros A, Szendroi A, Szucs M, Szekely E, Tokes AM, Romics I, Kulka J (2010) Beta-catenin expression and claudin expression pattern as prognostic factors of prostatic cancer progression. BJU Int 105:716–722
Tabaries S, Dong Z, Annis MG, Omeroglu A, Pepin F, Ouellet V, Russo C, Hassanain M, Metrakos P, Diaz Z, Basik M, Bertos N, Park M, Guettier C, Adam R, Hallett M, Siegel PM (2011) Claudin-2 is selectively enriched in and promotes the formation of breast cancer liver metastases through engagement of integrin complexes. Oncogene 30:1318–1328
Takahashi A, Komiya E, Kakutani H, Yoshida T, Fujii M, Horiguchi Y, Mizuguchi H, Tsutsumi Y, Tsunoda S, Koizumi N, Isoda K, Yagi K, Watanabe Y, Kondoh M (2008) Domain mapping of a claudin-4 modulator, the C-terminal region of C-terminal fragment of Clostridium perfringens enterotoxin, by site-directed mutagenesis. Biochem Pharmacol 75:1639–1648
Takahashi A, Kondoh M, Masuyama A, Fujii M, Mizuguchi H, Horiguchi Y, Watanabe Y (2005) Role of C-terminal regions of the C-terminal fragment of Clostridium perfringens enterotoxin in its interaction with claudin-4. J Control Release 108:56–62
Takahashi A, Kondoh M, Uchida H, Kakamu Y, Hamakubo T, Yagi K (2011) Mutated C-terminal fragments of Clostridium perfringens enterotoxin have increased affinity to claudin-4 and reversibly modulate tight junctions in vitro. Biochem Biophys Res Commun 410:466–470
Tassi RA, Bignotti E, Falchetti M, Ravanini M, Calza S, Ravaggi A, Bandiera E, Facchetti F, Pecorelli S, Santin AD (2008) Claudin-7 expression in human epithelial ovarian cancer. Int J Gynecol Cancer 18:1262–1271
Thuma F, Zoller M (2013) EpCAM-associated claudin-7 supports lymphatic spread and drug resistance in rat pancreatic cancer. Int J Cancer 133:855–866
Uchida H, Kondoh M, Hanada T, Takahashi A, Hamakubo T, Yagi K (2010) A claudin-4 modulator enhances the mucosal absorption of a biologically active peptide. Biochem Pharmacol 79:1437–1444
Ushiku T, Shinozaki-Ushiku A, Maeda D, Morita S, Fukayama M (2012) Distinct expression pattern of claudin-6, a primitive phenotypic tight junction molecule, in germ cell tumours and visceral carcinomas. Histopathology 61:1043–1056
Van Itallie CM, Betts L, Smedley JG 3rd, McClane BA, Anderson JM (2008) Structure of the claudin-binding domain of Clostridium perfringens enterotoxin. J Biol Chem 283:268–274
Veshnyakova A, Piontek J, Protze J, Waziri N, Heise I, Krause G (2012) Mechanism of Clostridium perfringens enterotoxin interaction with claudin-3/-4 protein suggests structural modifications of the toxin to target specific claudins. J Biol Chem 287:1698–1708
Wallace FM, Mach AS, Keller AM, Lindsay JA (1999) Evidence for Clostridium perfringens enterotoxin (CPE) inducing a mitogenic and cytokine response in vitro and a cytokine response in vivo. Curr Microbiol 38:96–100
Winkler L, Gehring C, Wenzel A, Muller SL, Piehl C, Krause G, Blasig IE, Piontek J (2009) Molecular determinants of the interaction between Clostridium perfringens enterotoxin fragments and claudin-3. J Biol Chem 284:18863–18872
Yoshida H, Sumi T, Zhi X, Yasui T, Honda KI, Ishiko O (2011) Claudin-4: a potential therapeutic target in chemotherapy-resistant ovarian cancer. Anticancer Res 31:1271–1277
Yuan X, Lin X, Manorek G, Kanatani I, Cheung LH, Rosenblum MG, Howell SB (2009) Recombinant CPE fused to tumor necrosis factor targets human ovarian cancer cells expressing the claudin-3 and claudin-4 receptors. Mol Cancer Ther 8:1906–1915
Zheng JY, Yu D, Foroohar M, Ko E, Chan J, Kim N, Chiu R, Pang S (2003) Regulation of the expression of the prostate-specific antigen by claudin-7. J Membrane Biol 194:187–197
Acknowledgments
We thank all of the members of our laboratory for their useful comments. This work was supported by a Health and Labour Sciences Research Grant from the Ministry of Health, Labour, and Welfare of Japan; a research grant from the Japan Agency for Medical Research and Development (AMED); a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan [grant number 24390042]; and funds from the Adaptable and Seamless Technology Transfer Program through Target-driven R&D Agency; Platform for Drug Discovery, Informatics, and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan; and the Takeda Science Foundation. Y.H. is supported by a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hashimoto, Y., Yagi, K. & Kondoh, M. Roles of the first-generation claudin binder, Clostridium perfringens enterotoxin, in the diagnosis and claudin-targeted treatment of epithelium-derived cancers. Pflugers Arch - Eur J Physiol 469, 45–53 (2017). https://doi.org/10.1007/s00424-016-1878-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-016-1878-6