Abstract
Purpose
This scoping review summarizes the applications of artificial intelligence (AI) and bioinformatics methodologies in analysis of ocular biofluid markers. The secondary objective was to explore supervised and unsupervised AI techniques and their predictive accuracies. We also evaluate the integration of bioinformatics with AI tools.
Methods
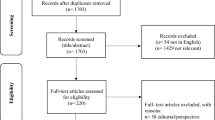
This scoping review was conducted across five electronic databases including EMBASE, Medline, Cochrane Central Register of Controlled Trials, Cochrane Database of Systematic Reviews, and Web of Science from inception to July 14, 2021. Studies pertaining to biofluid marker analysis using AI or bioinformatics were included.
Results
A total of 10,262 articles were retrieved from all databases and 177 studies met the inclusion criteria. The most commonly studied ocular diseases were diabetic eye diseases, with 50 papers (28%), while glaucoma was explored in 25 studies (14%), age-related macular degeneration in 20 (11%), dry eye disease in 10 (6%), and uveitis in 9 (5%). Supervised learning was used in 91 papers (51%), unsupervised AI in 83 (46%), and bioinformatics in 85 (48%). Ninety-eight papers (55%) used more than one class of AI (e.g. > 1 of supervised, unsupervised, bioinformatics, or statistical techniques), while 79 (45%) used only one. Supervised learning techniques were often used to predict disease status or prognosis, and demonstrated strong accuracy. Unsupervised AI algorithms were used to bolster the accuracy of other algorithms, identify molecularly distinct subgroups, or cluster cases into distinct subgroups that are useful for prediction of the disease course. Finally, bioinformatic tools were used to translate complex biomarker profiles or findings into interpretable data.
Conclusion
AI analysis of biofluid markers displayed diagnostic accuracy, provided insight into mechanisms of molecular etiologies, and had the ability to provide individualized targeted therapeutic treatment for patients. Given the progression of AI towards use in both research and the clinic, ophthalmologists should be broadly aware of the commonly used algorithms and their applications. Future research may be aimed at validating algorithms and integrating them in clinical practice.

Similar content being viewed by others
References
Rajkomar A, Dean J, Kohane I (2019) Machine Learning in Medicine. N Engl J Med 380:1347–1358. https://doi.org/10.1056/NEJMra1814259
Yu K-H, Beam AL, Kohane IS (2018) Artificial intelligence in healthcare. Nat Biomed Eng 2:719–731. https://doi.org/10.1038/s41551-018-0305-z
Pucchio A, Del Papa J, de Moraes FY (2022) Artificial intelligence in the medical profession: ready or not, here AI comes. Clinics 77:100010. https://doi.org/10.1016/j.clinsp.2022.100010
Tong Y, Lu W, Yu Y, Shen Y (2020) Application of machine learning in ophthalmic imaging modalities. Eye Vis 7:22. https://doi.org/10.1186/s40662-020-00183-6
Du XL, Li WB, Hu BJ (2018) Application of artificial intelligence in ophthalmology. Int J Ophthalmol 11:1555–1561. https://doi.org/10.18240/ijo.2018.09.21
Pomyen Y, Wanichthanarak K, Poungsombat P et al (2020) Deep metabolome: Applications of deep learning in metabolomics. Comput Struct Biotechnol J 18:2818–2825. https://doi.org/10.1016/j.csbj.2020.09.033
Csősz É, Tóth N, Deák E et al (2018) Wound-Healing Markers Revealed by Proximity Extension Assay in Tears of Patients following Glaucoma Surgery. Int J Mol Sci 19:4096. https://doi.org/10.3390/ijms19124096
Buisset A, Gohier P, Leruez S et al (2019) Metabolomic Profiling of Aqueous Humor in Glaucoma Points to Taurine and Spermine Deficiency: Findings from the Eye-D Study. J Proteome Res 18:1307–1315. https://doi.org/10.1021/acs.jproteome.8b00915
Han G, Wei P, He M et al (2020) Metabolomic Profiling of the Aqueous Humor in Patients with Wet Age-Related Macular Degeneration Using UHPLC-MS/MS. J Proteome Res 19:2358–2366. https://doi.org/10.1021/acs.jproteome.0c00036
Beutgen VM, Perumal N, Pfeiffer N, Grus FH (2019) Autoantibody Biomarker Discovery in Primary Open Angle Glaucoma Using Serological Proteome Analysis (SERPA). Front Immunol 10. https://doi.org/10.3389/fimmu.2019.00381
Sharma S, Bollinger KE, Kodeboyina SK et al (2018) Proteomic Alterations in Aqueous Humor From Patients With Primary Open Angle Glaucoma. Investig Opthalmology Vis Sci 59:2635. https://doi.org/10.1167/iovs.17-23434
Gao Y, Teo YCK, Beuerman RW et al (2020) A serum metabolomics study of patients with nAMD in response to anti-VEGF therapy. Sci Rep 10:1341. https://doi.org/10.1038/s41598-020-58346-3
Duong V-A, Ahn J, Han N-Y et al (2021) Proteomic Analysis of the Vitreous Body in Proliferative and Non-Proliferative Diabetic Retinopathy. Curr Proteomics 18:143–152. https://doi.org/10.2174/1570164617666200302101442
Li M, Li H, Jiang P et al (2014) Investigating the pathological processes of rhegmatogenous retinal detachment and proliferative vitreoretinopathy with metabolomics analysis. Mol Biosyst 10:1055. https://doi.org/10.1039/c3mb70386j
Anton Apreutesei N, Tircoveanu F, Cantemir A et al (2018) Predictions of ocular changes caused by diabetes in glaucoma patients. Comput Methods Programs Biomed 154:183–190. https://doi.org/10.1016/j.cmpb.2017.11.013
Adav SS, Wei J, Qian J et al (2019) Aqueous humor protein dysregulation in primary angle-closure glaucoma. Int Ophthalmol 39:861–871. https://doi.org/10.1007/s10792-018-0885-3
Iomdina EN, Tikhomirova NK, Bessmertny AM et al (2020) Alterations in proteome of human sclera associated with primary open-angle glaucoma involve proteins participating in regulation of the extracellular matrix. Mol Vis 26:623–640
Page MJ, McKenzie JE, Bossuyt PM et al (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372:n71. https://doi.org/10.1136/bmj.n71
Tricco AC, Lillie E, Zarin W et al (2018) PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann Intern Med 169:467–473. https://doi.org/10.7326/M18-0850
Grant MJ, Booth A (2009) A typology of reviews: an analysis of 14 review types and associated methodologies. Heal Inf Libr J 26:91–108. https://doi.org/10.1111/j.1471-1842.2009.00848.x
Alabdulwahhab KM, Sami W, Mehmood T et al (2021) Automated detection of diabetic retinopathy using machine learning classifiers. Eur Rev Med Pharmacol Sci 25:583–590. https://doi.org/10.26355/eurrev_202101_24615
Cao B, Zhang N, Zhang Y et al (2021) Plasma cytokines for predicting diabetic retinopathy among type 2 diabetic patients via machine learning algorithms. Aging 13:1972–1988. https://doi.org/10.18632/aging.202168
Barbosa Breda J, Croitor Sava A, Himmelreich U et al (2020) Metabolomic profiling of aqueous humor from glaucoma patients - The metabolomics in surgical ophthalmological patients (MISO) study. Exp Eye Res 201:108268. https://doi.org/10.1016/j.exer.2020.108268
Tang Y, Pan Y, Chen Y et al (2021) Metabolomic Profiling of Aqueous Humor and Plasma in Primary Open Angle Glaucoma Patients Points Towards Novel Diagnostic and Therapeutic Strategy. Front Pharmacol 12:1–12. https://doi.org/10.3389/fphar.2021.621146
Curnow SJ, Falciani F, Durrani OM et al (2005) Multiplex Bead Immunoassay Analysis of Aqueous Humor Reveals Distinct Cytokine Profiles In Uveitis. Investig Opthalmology Vis Sci 46:4251. https://doi.org/10.1167/iovs.05-0444
Igarashi N, Honjo M, Asaoka R et al (2021) Aqueous autotaxin and TGF-βs are promising diagnostic biomarkers for distinguishing open-angle glaucoma subtypes. Sci Rep 11:1408. https://doi.org/10.1038/s41598-021-81048-3
Lynch AM, Wagner BD, Palestine AG et al (2020) Plasma Biomarkers of Reticular Pseudodrusen and the Risk of Progression to Advanced Age-Related Macular Degeneration. Transl Vis Sci Technol 9:12. https://doi.org/10.1167/tvst.9.10.12
Testa MA, Puklin JE, Sherwin RS, Simonson DC (1985) Clinical Predictors of Retinopathy and Its Progression in Patients with Type I Diabetes During CSII or Conventional Insulin Treatment. Diabetes 34:61–68. https://doi.org/10.2337/diab.34.3.S61
de Almeida BD, Alborghetti MR, Franco Paes Leme A et al (2020) Tear proteomic profile in three distinct ocular surface diseases: keratoconus, pterygium, and dry eye related to graft-versus-host disease. Clin Proteomics 17:42. https://doi.org/10.1186/s12014-020-09307-5
González N, Iloro I, Soria J et al (2014) Human tear peptide/protein profiling study of ocular surface diseases by SPE-MALDI-TOF mass spectrometry analyses. EuPA Open Proteom 3:206–215. https://doi.org/10.1016/j.euprot.2014.02.016
Grus FH, Joachim SC, Sandmann S et al (2008) Transthyretin and complex protein pattern in aqueous humor of patients with primary open-angle glaucoma. Mol Vis 14:1437–1445
Hysi PG, Khawaja AP, Menni C et al (2019) Ascorbic acid metabolites are involved in intraocular pressure control in the general population. Redox Biol 20:349–353. https://doi.org/10.1016/j.redox.2018.10.004
Ji DY, Park SH, Park SJ et al (2018) Comparative assessment of Graves’ disease and main extrathyroidal manifestation, Graves’ ophthalmopathy, by non-targeted metabolite profiling of blood and orbital tissue. Sci Rep 8:9262. https://doi.org/10.1038/s41598-018-27600-0
Xu J, Su G, Huang X et al (2021) Metabolomic Analysis of Aqueous Humor Identifies Aberrant Amino Acid and Fatty Acid Metabolism in Vogt-Koyanagi-Harada and Behcet’s Disease. Front Immunol 12:1–10. https://doi.org/10.3389/fimmu.2021.587393
Barba I, Garcia-Ramírez M, Hernández C et al (2010) Metabolic Fingerprints of Proliferative Diabetic Retinopathy: An 1 H-NMR–Based Metabonomic Approach Using Vitreous Humor. Investig Opthalmology Vis Sci 51:4416. https://doi.org/10.1167/iovs.10-5348
Blighe K, Gurudas S, Lee Y, Sivaprasad S (2020) Diabetic Retinopathy Environment-Wide Association Study (EWAS) in NHANES 2005–2008. J Clin Med 9:3643. https://doi.org/10.3390/jcm9113643
Bocca C, Kouassi Nzoughet J, Leruez S et al (2018) A Plasma Metabolomic Signature Involving Purine Metabolism in Human Optic Atrophy 1 ( OPA1)-Related Disorders. Investig Opthalmology Vis Sci 59:185. https://doi.org/10.1167/iovs.17-23027
Bocca C, Le Paih V, Chao de la Barca JM et al (2021) A plasma metabolomic signature of Leber hereditary optic neuropathy showing taurine and nicotinamide deficiencies. Hum Mol Genet 30:21–29. https://doi.org/10.1093/hmg/ddab013
Han G, Wei P, He M et al (2020) Metabolomic Profiling of the Aqueous Humor in Patients with Wet Age-Related Macular Degeneration Using UHPLC–MS/MS. J Proteome Res 19:2358–2366. https://doi.org/10.1021/acs.jproteome.0c00036
Osborn MP, Park Y, Parks MB et al (2013) Metabolome-Wide Association Study of Neovascular Age-Related Macular Degeneration. PLoS One 8:e72737. https://doi.org/10.1371/journal.pone.0072737
Shimizu H, Usui Y, Wakita R et al (2021) Differential Tissue Metabolic Signatures in IgG4-Related Ophthalmic Disease and Orbital Mucosa-Associated Lymphoid Tissue Lymphoma. Investig Opthalmology Vis Sci 62:15. https://doi.org/10.1167/iovs.62.1.15
Soria J, Villarrubia A, Merayo-Lloves J et al (2015) Label-free LC-MS/MS quantitative analysis of aqueous humor from keratoconic and normal eyes. Mol Vis 21:451–460
Verhagen FH, Stigter ECA, Pras-Raves ML et al (2019) Aqueous Humor Analysis Identifies Higher Branched Chain Amino Acid Metabolism as a Marker for Human Leukocyte Antigen-B27 Acute Anterior Uveitis and Disease Activity. Am J Ophthalmol 198:97–110. https://doi.org/10.1016/j.ajo.2018.10.004
Wojakowska A, Pietrowska M, Widlak P et al (2020) Metabolomic Signature Discriminates Normal Human Cornea from Keratoconus—A Pilot GC/MS Study. Molecules 25:2933. https://doi.org/10.3390/molecules25122933
Mitchell SL, Uppal K, Williamson SM et al (2018) The Carnitine Shuttle Pathway is Altered in Patients With Neovascular Age-Related Macular Degeneration. Investig Opthalmology Vis Sci 59:4978. https://doi.org/10.1167/iovs.18-25137
Luo D, Deng T, Yuan W et al (2017) Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol 17:165. https://doi.org/10.1186/s12886-017-0555-7
Zhavoronkov A, Izumchenko E, Kanherkar RR et al (2016) Pro-fibrotic pathway activation in trabecular meshwork and lamina cribrosa is the main driving force of glaucoma. Cell Cycle 15:1643–1652. https://doi.org/10.1080/15384101.2016.1170261
Bonacini M, Soriano A, Cimino L et al (2020) Cytokine Profiling in Aqueous Humor Samples From Patients With Non-Infectious Uveitis Associated With Systemic Inflammatory Diseases. Front Immunol 11:1–12. https://doi.org/10.3389/fimmu.2020.00358
Jiang L, Rong A, Wei R et al (2020) Tear proteomics of orbital decompression for disfiguring exophthalmos in inactive thyroid-associated ophthalmopathy. Exp Ther Med 20:1–1. https://doi.org/10.3892/etm.2020.9383
Sembler-Møller ML, Belstrøm D, Locht H, Pedersen AML (2020) Proteomics of saliva, plasma, and salivary gland tissue in Sjögren’s syndrome and non-Sjögren patients identify novel biomarker candidates. J Proteomics 225:103877. https://doi.org/10.1016/j.jprot.2020.103877
Bancks MP, Casanova R, Gregg EW, Bertoni AG (2019) Epidemiology of diabetes phenotypes and prevalent cardiovascular risk factors and diabetes complications in the National Health and Nutrition Examination Survey 2003–2014. Diabetes Res Clin Pract 158:107915. https://doi.org/10.1016/j.diabres.2019.107915
Karpati T, Leventer-Roberts M, Feldman B et al (2018) Patient clusters based on HbA1c trajectories: A step toward individualized medicine in type 2 diabetes. PLoS One 13:e0207096. https://doi.org/10.1371/journal.pone.0207096
Aretz S, Krohne TU, Kammerer K et al (2013) In-depth mass spectrometric mapping of the human vitreous proteome. Proteome Sci 11:22. https://doi.org/10.1186/1477-5956-11-22
Dor M, Eperon S, Lalive PH et al (2019) Investigation of the global protein content from healthy human tears. Exp Eye Res 179:64–74. https://doi.org/10.1016/j.exer.2018.10.006
Velez G, Bassuk AG, Colgan D et al (2017) Therapeutic drug repositioning using personalized proteomics of liquid biopsies. JCI Insight 2. https://doi.org/10.1172/jci.insight.97818
Lee S, Zhou J, Wong WT et al (2021) Glycemic and lipid variability for predicting complications and mortality in diabetes mellitus using machine learning. BMC Endocr Disord 21:94. https://doi.org/10.1186/s12902-021-00751-4
Yao L, Zhong Y, Wu J et al (2019) Multivariable Logistic Regression And Back Propagation Artificial Neural Network To Predict Diabetic Retinopathy. Diabetes, Metab Syndr Obes Targets Ther 12:1943–1951. https://doi.org/10.2147/DMSO.S219842
Marino APMP, dos Santos LI, Henriques PM et al (2020) Circulating inflammatory mediators as biomarkers of ocular toxoplasmosis in acute and in chronic infection. J Leukoc Biol 108:1253–1264. https://doi.org/10.1002/JLB.4MA0420-702R
Liu B, Zhang B, Hu Y et al (2021) Automatic prediction of treatment outcomes in patients with diabetic macular edema using ensemble machine learning. Ann Transl Med 9:43–43. https://doi.org/10.21037/atm-20-1431
Mazumder AG, Banerjee S, Zevictovich F et al (2018) Fourier-transform-infrared-spectroscopy based metabolomic spectral biomarker selection towards optimal diagnostic differentiation of diabetes with and without retinopathy. Spectrosc Lett 51:340–349. https://doi.org/10.1080/00387010.2018.1471510
Schori C, Trachsel C, Grossmann J et al (2018) The proteomic landscape in the vitreous of patients with age-related and diabetic retinal disease. Investig Ophthalmol Vis Sci 59:AMD31–AMD40. https://doi.org/10.1167/iovs.18-24122
Torok Z, Peto T, Csosz E et al (2013) Tear fluid proteomics multimarkers for diabetic retinopathy screening. BMC Ophthalmol 13:40. https://doi.org/10.1186/1471-2415-13-40
Li M, Zhang X, Liao N et al (2016) Analysis of the Serum Lipid Profile in Polypoidal Choroidal Vasculopathy. Sci Rep 6:38342. https://doi.org/10.1038/srep38342
Pucchio A, Krance SH, Pur DR et al (2022) Artificial Intelligence Analysis of Biofluid Markers in Age-Related Macular Degeneration: A Systematic Review. Clin Ophthalmol 16:2463–2476. https://doi.org/10.2147/OPTH.S377262
Pucchio A, Krance S, Pur DR et al (2022) The role of artificial intelligence in analysis of biofluid markers for diagnosis and management of glaucoma: A systematic review. Eur J Ophthalmol 112067212211409. https://doi.org/10.1177/11206721221140948
Pur DR, Krance SH, Pucchio A et al (2022) Current uses of artificial intelligence in the analysis of biofluid markers involved in corneal and ocular surface diseases: a systematic review. Eye 1–3. https://doi.org/10.1038/s41433-022-02307-9
Bassi A, Krance SH, Pucchio A et al (2022) The Application of Artificial Intelligence in the Analysis of Biomarkers for Diagnosis and Management of Uveitis and Uveal Melanoma: A Systematic Review. Clin Ophthalmol 16:2895–2908. https://doi.org/10.2147/OPTH.S377358
Pur DR, Krance S, Pucchio A et al (2022) Emerging applications of bioinformatics and artificial intelligence in the analysis of biofluid markers involved in retinal occlusive diseases: a systematic review. Graefe’s Arch Clin Exp Ophthalmol. https://doi.org/10.1007/s00417-022-05769-5
Maulud D, Abdulazeez AM (2020) A Review on Linear Regression Comprehensive in Machine Learning. J Appl Sci Technol Trends 1:140–147. https://doi.org/10.38094/jastt1457
Ing EB, Miller NR, Nguyen A et al (2019) Neural network and logistic regression diagnostic prediction models for giant cell arteritis: development and validation. Clin Ophthalmol 13:421–430. https://doi.org/10.2147/OPTH.S193460
Burgess LG, Uppal K, Walker DI et al (2015) Metabolome-Wide Association Study of Primary Open Angle Glaucoma. Investig Opthalmology Vis Sci 56:5020. https://doi.org/10.1167/iovs.15-16702
Tokuda Y, Yagi T, Yoshii K et al (2012) An approach to predict the risk of glaucoma development by integrating different attribute data. Springerplus 1:41. https://doi.org/10.1186/2193-1801-1-41
Aumüller M, Bernhardsson E, Faithfull A (2020) ANN-Benchmarks: A benchmarking tool for approximate nearest neighbor algorithms. Inf Syst 87:101374. https://doi.org/10.1016/j.is.2019.02.006
Pisner DA, Schnyer DM (2020) Support vector machine. In: Machine Learning. Elsevier, pp 101–121 More details here: https://www.sciencedirect.com/science/article/abs/pii/B9780128157398000067
Zou J, Han Y, So S-S (2008) Overview of artificial neural networks. pp 14–22 More details here: https://pubmed.ncbi.nlm.nih.gov/19065803/
Hengl T, Nussbaum M, Wright MN et al (2018) Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 6:e5518. https://doi.org/10.7717/peerj.5518
North Carolina State University (2022) PA 765: Discriminant Function Analysis. In: Web Arch. https://web.archive.org/web/20080312065328/http://www2.chass.ncsu.edu/garson/pA765/discrim.htm. Accessed 21 Sep 2022
Ao Y, Li H, Zhu L et al (2019) The linear random forest algorithm and its advantages in machine learning assisted logging regression modeling. J Pet Sci Eng 174:776–789. https://doi.org/10.1016/j.petrol.2018.11.067
Ayinla S, Kehinde Adekunle B. An Overview and Application of Discriminant Analysis in Data Analysis. IOSR J Math 11:2278–5728. https://doi.org/10.9790/5728-11151215
Karamizadeh S, Abdullah SM, Manaf AA et al (2013) An Overview of Principal Component Analysis. J Signal Inf Process 04:173–175. https://doi.org/10.4236/jsip.2013.43B031
Acar İE, Lores-Motta L, Colijn JM, et al (2020) Integrating metabolomics, genomics, and disease pathways in age-related macular degeneration: The EYE-RISK Consortium More details here: https://pubmed.ncbi.nlm.nih.gov/32553749/
Agasing AM, Wu Q, Khatri B et al (2020) Transcriptomics and proteomics reveal a cooperation between interferon and T-helper 17 cells in neuromyelitis optica. Nat Commun 11:2856. https://doi.org/10.1038/s41467-020-16625-7
Agrawal R, Grant R, Gupta B et al (2017) What does IGRA testing add to the diagnosis of ocular tuberculosis? A Bayesian latent class analysis. BMC Ophthalmol 17:245. https://doi.org/10.1186/s12886-017-0597-x
Ahlqvist E, Storm P, Käräjämäki A et al (2018) Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol 6:361–369. https://doi.org/10.1016/S2213-8587(18)30051-2
Anjana RM, Baskar V, Nair ATN et al (2020) Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data-driven cluster analysis: the INSPIRED study. BMJ open diabetes Res care 8:e001506. https://doi.org/10.1136/bmjdrc-2020-001506
Aqrawi LA, Galtung HK, Vestad B et al (2017) Identification of potential saliva and tear biomarkers in primary Sjögren’s syndrome, utilising the extraction of extracellular vesicles and proteomics analysis. Arthritis Res Ther 19:14. https://doi.org/10.1186/s13075-017-1228-x
Bai S, Liu S, Guo X et al (2010) Proteome analysis of haptoglobin in cerebrospinal fluid of neuromyelitis optica. Mol Biol Rep 37:1619–1625. https://doi.org/10.1007/s11033-009-9574-7
Bennett KL, Funk M, Tschernutter M et al (2011) Proteomic analysis of human cataract aqueous humour: Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ®-labelled specimens. J Proteomics 74:151–166. https://doi.org/10.1016/j.jprot.2010.10.002
Beutgen VM, Pfeiffer N, Grus FH (2021) Serological Levels of Anti-clathrin Antibodies Are Decreased in Patients With Pseudoexfoliation Glaucoma. Front Immunol 12:1–9. https://doi.org/10.3389/fimmu.2021.616421
Breda JB, Sava AC, Himmelreich U et al (2020) Metabolomic profiling of aqueous humor from glaucoma patients - The metabolomics in surgical ophthalmological patients (MISO) study. Exp Eye Res 201:108268. https://doi.org/10.1016/j.exer.2020.108268
Chiang SY, Tsai ML, Wang CY et al (2012) Proteomic analysis and identification of aqueous humor proteins with a pathophysiological role in diabetic retinopathy. J Proteomics 75:2950–2959. https://doi.org/10.1016/j.jprot.2011.12.006
Christakopoulos C, Cehofski LJ, Christensen SR et al (2019) Proteomics reveals a set of highly enriched proteins in epiretinal membrane compared with inner limiting membrane. Exp Eye Res 186:107722. https://doi.org/10.1016/j.exer.2019.107722
Curovic VR, Suvitaival T, Mattila I et al (2020) Circulating Metabolites and Lipids Are Associated to Diabetic Retinopathy in Individuals With Type 1 Diabetes. Diabetes 69:2217–2226. https://doi.org/10.2337/db20-0104
Dagliati A, Marini S, Sacchi L et al (2018) Machine Learning Methods to Predict Diabetes Complications. J Diabetes Sci Technol 12:295–302. https://doi.org/10.1177/1932296817706375
Fan Y, Long E, Cai L et al (2021) Machine Learning Approaches to Predict Risks of Diabetic Complications and Poor Glycemic Control in Nonadherent Type 2 Diabetes. Front Pharmacol 12:1–11. https://doi.org/10.3389/fphar.2021.665951
Fernández I, López-Miguel A, Enríquez-de-Salamanca A et al (2019) Response profiles to a controlled adverse desiccating environment based on clinical and tear molecule changes. Ocul Surf 17:502–515. https://doi.org/10.1016/j.jtos.2019.03.009
Fernández I, López-Miguel A, Martín-Montañez V et al (2020) Inflammatory status predicts contact lens discomfort under adverse environmental conditions. Ocul Surf 18:829–840. https://doi.org/10.1016/j.jtos.2020.07.015
Grus FH, Augustin AJ, Toth-Sagi K (1998) Diagnostic classification of autoantibody repertoires in endocrine ophthalmopathy using an artificial neural network. Ocul Immunol Inflamm 6:43–50. https://doi.org/10.1076/ocii.6.1.43.8082
Grus FH, Sabuncuo P, Augustin AJ (2001) Analysis of tear protein patterns of dry-eye patients using fluorescent staining dyes and two-dimensional quantification algorithms. Electrophoresis 22:1845–1850. https://doi.org/10.1002/1522-2683(200105)22:9%3c1845::AID-ELPS1845%3e3.0.CO;2-N
Grus FH, Kramann C, Bozkurt N et al (2005) Effects of multipurpose contact lens solutions on the protein composition of the tear film. Contact Lens Anterior Eye 28:103–112. https://doi.org/10.1016/j.clae.2005.06.004
Grus FH, Podust VN, Bruns K et al (2005) SELDI-TOF-MS ProteinChip Array Profiling of Tears from Patients with Dry Eye. Investig Opthalmology Vis Sci 46:863. https://doi.org/10.1167/iovs.04-0448
Guha Mazumder A, Chatterjee S, Chatterjee S et al (2017) Spectropathology-corroborated multimodal quantitative imaging biomarkers for neuroretinal degeneration in diabetic retinopathy. Clin Ophthalmol 11:2073–2089. https://doi.org/10.2147/OPTH.S140110
Guo J, Yan T, Bi H et al (2014) Plasma metabonomics study of the patients with acute anterior uveitis based on ultra-performance liquid chromatography–mass spectrometry. Graefe’s Arch Clin Exp Ophthalmol 252:925–934. https://doi.org/10.1007/s00417-014-2619-1
Hall JC, Baer AN, Shah AA et al (2015) Molecular Subsetting of Interferon Pathways in Sjögren’s Syndrome. Arthritis Rheumatol 67:2437–2446. https://doi.org/10.1002/art.39204
Huang Z, Du C-X, Pan X-D (2018) The use of in-strip digestion for fast proteomic analysis on tear fluid from dry eye patients. PLoS One 13:e0200702. https://doi.org/10.1371/journal.pone.0200702
Indini A, Di Guardo L, Cimminiello C et al (2019) Artificial Intelligence Estimates the Importance of Baseline Factors in Predicting Response to Anti-PD1 in Metastatic Melanoma. Am J Clin Oncol 42:643–648. https://doi.org/10.1097/COC.0000000000000566
Ji Y, Rong X, Ye H et al (2015) Proteomic analysis of aqueous humor proteins associated with cataract development. Clin Biochem 48:1304–1309. https://doi.org/10.1016/j.clinbiochem.2015.08.006
Ji YW, Kim HM, Ryu SY et al (2019) Changes in Human Tear Proteome Following Topical Treatment of Dry Eye Disease: Cyclosporine A Versus Diquafosol Tetrasodium. Investig Opthalmology Vis Sci 60:5035. https://doi.org/10.1167/iovs.19-27872
Jin H, Zhu B, Liu X et al (2019) Metabolic characterization of diabetic retinopathy: An 1H-NMR-based metabolomic approach using human aqueous humor. J Pharm Biomed Anal 174:414–421. https://doi.org/10.1016/j.jpba.2019.06.013
Joachim SC, Pfeiffer N, Grus FH (2005) Autoantibodies in patients with glaucoma: a comparison of IgG serum antibodies against retinal, optic nerve, and optic nerve head antigens. Graefe’s Arch Clin Exp Ophthalmol 243:817–823. https://doi.org/10.1007/s00417-004-1094-5
Joachim SC, Wuenschig D, Pfeiffer N, Grus FH (2007) IgG antibody patterns in aqueous humor of patients with primary open angle glaucoma and pseudoexfoliation glaucoma. Mol Vis 13:1573–1579
Jurynczyk M, Probert F, Yeo T et al (2017) Metabolomics reveals distinct, antibody-independent, molecular signatures of MS, AQP4-antibody and MOG-antibody disease. Acta Neuropathol Commun 5:95. https://doi.org/10.1186/s40478-017-0495-8
Kahkoska AR, Nguyen CT, Adair LA et al (2019) Longitudinal Phenotypes of Type 1 Diabetes in Youth Based on Weight and Glycemia and Their Association With Complications. J Clin Endocrinol Metab 104:6003–6016. https://doi.org/10.1210/jc.2019-00734
Ke C, Xu H, Chen Q et al (2021) Serum metabolic signatures of high myopia among older Chinese adults. Eye 35:817–824. https://doi.org/10.1038/s41433-020-0968-z
Kersten E, Dammeier S, Ajana S et al (2019) Metabolomics in serum of patients with non-advanced age-related macular degeneration reveals aberrations in the glutamine pathway. PLoS One 14:e0218457. https://doi.org/10.1371/journal.pone.0218457
Kim WB, Chung HK, Park YJ et al (2001) Clinical Significance of Classification of Graves` Disease According to the Characteristics of TSH receptor Antibodies. Korean J Intern Med 16:187–200. https://doi.org/10.3904/kjim.2001.16.3.187
Kim SW, Lee J, Lee B, Rhim T (2014) Proteomic analysis in pterygium; upregulated protein expression of ALDH3A1, PDIA3, and PRDX2. Mol Vis 20:1192–1202
Kouassi Nzoughet J, Guehlouz K, Leruez S et al (2020) A Data Mining Metabolomics Exploration of Glaucoma. Metabolites 10:49. https://doi.org/10.3390/metabo10020049
Kowalczuk L, Matet A, Dor M et al (2018) Proteome and Metabolome of Subretinal Fluid in Central Serous Chorioretinopathy and Rhegmatogenous Retinal Detachment: A Pilot Case Study. Transl Vis Sci Technol 7:3. https://doi.org/10.1167/tvst.7.1.3
Kramann C, Boehm N, Lorenz K et al (2011) Effect of contact lenses on the protein composition in tear film: a ProteinChip study. Graefe’s Arch Clin Exp Ophthalmol 249:233–243. https://doi.org/10.1007/s00417-010-1456-0
Kuiper JJW, Beretta L, Nierkens S et al (2017) An Ocular Protein Triad Can Classify Four Complex Retinal Diseases. Sci Rep 7:1–9. https://doi.org/10.1038/srep41595
Laíns I, Kelly RS, Miller JB et al (2018) Human Plasma Metabolomics Study across All Stages of Age-Related Macular Degeneration Identifies Potential Lipid Biomarkers. Ophthalmology 125:245–254. https://doi.org/10.1016/j.ophtha.2017.08.008
Laíns I, Chung W, Kelly RS et al (2019) Human Plasma Metabolomics in Age-Related Macular Degeneration: Meta-Analysis of Two Cohorts. Metabolites 9:127. https://doi.org/10.3390/metabo9070127
Li H, Ma C, Liu W et al (2020) Gypenosides Protect Orbital Fibroblasts in Graves Ophthalmopathy via Anti-Inflammation and Anti-Fibrosis Effects. Investig Opthalmology Vis Sci 61:64. https://doi.org/10.1167/iovs.61.5.64
Li J, Wang T, Zuo J et al (2020) Association of n-6 PUFAs with the risk of diabetic retinopathy in diabetic patients. Endocr Connect 9:1191–1201. https://doi.org/10.1530/EC-20-0370
Linghu D, Guo L, Zhao Y et al (2017) iTRAQ-based quantitative proteomic analysis and bioinformatics study of proteins in pterygia. PROTEOMICS - Clin Appl 11:1600094. https://doi.org/10.1002/prca.201600094
Liu H, Zhang J, Zhou P et al (2019) Exploration of vascular adhesion protein-1 expression in patients with conjunctivitis associated systemic lupus erythematosus using 2D-DIGE. Exp Ther Med 5072–5077. https://doi.org/10.3892/etm.2019.8009
Liu A, Wang L, Feng Q et al (2021) Low expression of GSTP1 in the aqueous humour of patients with primary open-angle glaucoma. J Cell Mol Med 25:3063–3079. https://doi.org/10.1111/jcmm.16361
Liu Y-C, Yam GH-F, Lin MT-Y et al (2021) Comparison of tear proteomic and neuromediator profiles changes between small incision lenticule extraction (SMILE) and femtosecond laser-assisted in-situ keratomileusis (LASIK). J Adv Res 29:67–81. https://doi.org/10.1016/j.jare.2020.11.001
Lynch AM, Wagner BD, Weiss SJ et al (2019) Proteomic profiles in advanced age-related macular degeneration using an aptamer-based proteomic technology. Transl Vis Sci Technol 8. https://doi.org/10.1167/tvst.8.1.14
Lynch AM, Wagner BD, Palestine AG et al (2020) Plasma biomarkers of reticular pseudodrusen and the risk of progression to advanced age-related macular degeneration. Transl Vis Sci Technol 9:1–13. https://doi.org/10.1167/tvst.9.10.12
Maier P, Heizmann U, Böhringer D et al (2011) Predicting the risk for corneal graft rejection by aqueous humor analysis. Mol Vis 17:1016–1023
Menegay M, Lee D, Tabbara KF et al (2008) Proteomic Analysis of Climatic Keratopathy Droplets. Investig Opthalmology Vis Sci 49:2829. https://doi.org/10.1167/iovs.07-1438
Mirić D, Kisić B, Žorić L et al (2012) The Impact of Senile Cataract Maturity on Blood Oxidative Stress Markers and Glutathione-Dependent Antioxidants: Relations with Lens Variables. J Med Biochem 31:184–192. https://doi.org/10.2478/v10011-012-0003-4
Moussallieh F-M, Elbayed K, Chanson J et al (2014) Serum analysis by 1 H Nuclear Magnetic Resonance spectroscopy: a new tool for distinguishing neuromyelitis optica from multiple sclerosis. Mult Scler J 20:558–565. https://doi.org/10.1177/1352458513504638
Myer C, Abdelrahman L, Banerjee S et al (2020) Aqueous humor metabolite profile of pseudoexfoliation glaucoma is distinctive. Mol Omi 16:425–435. https://doi.org/10.1039/C9MO00192A
Nättinen J, Mäkinen P, Aapola U et al (2020) Early changes in tear film protein profiles after femtosecond LASIK surgery. Clin Proteomics 17:36. https://doi.org/10.1186/s12014-020-09303-9
Nezu N, Usui Y, Saito A et al (2021) Machine Learning Approach for Intraocular Disease Prediction Based on Aqueous Humor Immune Mediator Profiles. Ophthalmology 128:1197–1208. https://doi.org/10.1016/j.ophtha.2021.01.019
Nielsen HH, Beck HC, Kristensen LP et al (2015) The Urine Proteome Profile Is Different in Neuromyelitis Optica Compared to Multiple Sclerosis: A Clinical Proteome Study. PLoS One 10:e0139659. https://doi.org/10.1371/journal.pone.0139659
Nusinovici S, Zhang L, Chai X, et al (2020) Machine learning to determine relative contribution of modifiable and non-modifiable risk factors of major eye diseases. Br J Ophthalmol 2020–317454. https://doi.org/10.1136/bjophthalmol-2020-317454
Ogunyemi OI, Gandhi M, Tayek C (2019) Predictive Models for Diabetic Retinopathy from Non-Image Teleretinal Screening Data. AMIA Jt Summits Transl Sci proceedings AMIA Jt Summits Transl Sci 2019:472–477
Okrojek R, Grus FH, Matheis N, Kahaly GJ (2009) Proteomics in Autoimmune Thyroid Eye Disease. Horm Metab Res 41:465–470. https://doi.org/10.1055/s-0029-1214413
O’Leary OE, Schoetzau A, Amruthalingam L et al (2020) Tear Proteomic Predictive Biomarker Model for Ocular Graft Versus Host Disease Classification. Transl Vis Sci Technol 9:3. https://doi.org/10.1167/tvst.9.9.3
Pan C-W, Ke C, Chen Q et al (2020) Differential metabolic markers associated with primary open-angle glaucoma and cataract in human aqueous humor. BMC Ophthalmol 20:183. https://doi.org/10.1186/s12886-020-01452-7
Pavan J, Štambuk N, Pokrić B et al (2000) Quantification of Intraocular Interferon-γ and IgG in Cataract and Diabetes. Croat Chem Acta 73:1099–1110
Prior MJ, Prout T, Miller D et al (1993) C-peptide and the classification of diabetes mellitus patients in the Early treatment diabetic retinopathy study report number 6. Ann Epidemiol 3:9–17. https://doi.org/10.1016/1047-2797(93)90004-N
Qin Y, Feng X, Luo H et al (2022) Association between plasma free fatty acid levels and primary angle-closure glaucoma based on a mass spectrometry metabolomics analysis. Acta Ophthalmol 100:e204–e212. https://doi.org/10.1111/aos.14874
Romero P, Salvat M, Fernández J et al (2007) Renal and retinal microangiopathy after 15 years of follow-up study in a sample of Type 1 diabetes mellitus patients. J Diabetes Complications 21:93–100. https://doi.org/10.1016/j.jdiacomp.2006.04.001
Romero-Aroca P, Baget-Bernaldiz M, Reyes-Torres J et al (2012) Relationship between diabetic retinopathy, microalbuminuria and overt nephropathy, and twenty-year incidence follow-up of a sample of type 1 diabetic patients. J Diabetes Complications 26:506–512. https://doi.org/10.1016/j.jdiacomp.2012.06.010
Romero-Aroca P, Valls A, Moreno A et al (2019) A Clinical Decision Support System for Diabetic Retinopathy Screening: Creating a Clinical Support Application. Telemed e-Health 25:31–40. https://doi.org/10.1089/tmj.2017.0282
Romero-Aroca P, Verges-Pujol R, Santos-Blanco E et al (2021) Validation of a Diagnostic Support System for Diabetic Retinopathy Based on Clinical Parameters. Transl Vis Sci Technol 10:17. https://doi.org/10.1167/tvst.10.3.17
Roybal CN, Velez G, Toral MA et al (2018) Personalized Proteomics in Proliferative Vitreoretinopathy Implicate Hematopoietic Cell Recruitment and mTOR as a Therapeutic Target. Am J Ophthalmol 186:152–163. https://doi.org/10.1016/j.ajo.2017.11.025
Safai N, Ali A, Rossing P, Ridderstråle M (2018) Stratification of type 2 diabetes based on routine clinical markers. Diabetes Res Clin Pract 141:275–283. https://doi.org/10.1016/j.diabres.2018.05.014
Sandhu HS, Elmogy M, Taher Sharafeldeen A et al (2020) Automated Diagnosis of Diabetic Retinopathy Using Clinical Biomarkers, Optical Coherence Tomography, and Optical Coherence Tomography Angiography. Am J Ophthalmol 216:201–206. https://doi.org/10.1016/j.ajo.2020.01.016
Sato T, Takeuchi M, Karasawa Y et al (2019) Comprehensive expression patterns of inflammatory cytokines in aqueous humor of patients with neovascular age-related macular degeneration. Sci Rep 9:19447. https://doi.org/10.1038/s41598-019-55191-x
Seghieri G, Bartolomei G, De Giorgio LA (1986) Plasma fibronectin in diabetic retinopathy and macroangiopathy. Diabete Metab 12:186–90
Semeraro F, Parrinello G, Cancarini A et al (2011) Predicting the risk of diabetic retinopathy in type 2 diabetic patients. J Diabetes Complications 25:292–297. https://doi.org/10.1016/j.jdiacomp.2010.12.002
Shi XY, Li Q, Bin Wei W, Tao LM (2017) Peptidome profiling of human serum of uveal melanoma patients based on magnetic bead fractionation and mass spectrometry. Int J Ophthalmol 10:939–947. https://doi.org/10.18240/ijo.2017.06.17
Shimizu H, Usui Y, Asakage M et al (2020) Serum metabolomic profiling of patients with non-infectious uveitis. J Clin Med 9:1–10. https://doi.org/10.3390/jcm9123955
Sivagurunathan S, Selvan LN, Khan A et al (2021) Proteomics-based approach for differentiation of age-related macular degeneration sub-types. Indian J Ophthalmol 69:647. https://doi.org/10.4103/ijo.IJO_470_20
Skeie JM, Roybal CN, Mahajan VB (2015) Proteomic Insight into the Molecular Function of the Vitreous. PLoS One 10:e0127567. https://doi.org/10.1371/journal.pone.0127567
Sommer F, Spörl E, Herber R et al (2019) Predictive value of positive temporal artery biopsies in patients with clinically suspected giant cell arteritis considering temporal artery ultrasound findings. Graefe’s Arch Clin Exp Ophthalmol 257:2279–2284. https://doi.org/10.1007/s00417-019-04430-y
Srinivasan S, Thangavelu M, Zhang L et al (2012) iTRAQ Quantitative Proteomics in the Analysis of Tears in Dry Eye Patients. Investig Opthalmology Vis Sci 53:5052. https://doi.org/10.1167/iovs.11-9022
Sudha D, Kohansal-Nodehi M, Kovuri P et al (2017) Proteomic profiling of human intraschisis cavity fluid. Clin Proteomics 14:13. https://doi.org/10.1186/s12014-017-9148-y
Tebani A, Mauhin W, Abily-Donval L et al (2020) A Proteomics-Based Analysis Reveals Predictive Biological Patterns in Fabry Disease. J Clin Med 9:1325. https://doi.org/10.3390/jcm9051325
Ting DSW, Cheung CY, Nguyen Q et al (2019) Deep learning in estimating prevalence and systemic risk factors for diabetic retinopathy: a multi-ethnic study. NPJ Digit Med 2:24. https://doi.org/10.1038/s41746-019-0097-x
Tong L, Zhou L, Beuerman R et al (2017) Effects of punctal occlusion on global tear proteins in patients with dry eye. Ocul Surf 15:736–741. https://doi.org/10.1016/j.jtos.2017.04.002
Torok Z, Peto T, Csosz E et al (2015) Combined Methods for Diabetic Retinopathy Screening, Using Retina Photographs and Tear Fluid Proteomics Biomarkers. J Diabetes Res 2015:1–8. https://doi.org/10.1155/2015/623619
Tsubota K, Usui Y, Goto H (2020) Identification of Prognostic Markers in Patients with Primary Vitreoretinal Lymphoma by Clustering Analysis Using Clinical Data. J Clin Med 9:2298. https://doi.org/10.3390/jcm9072298
Tsubota K, Usui Y, Nemoto R, Goto H (2020) Identification of Markers Predicting Clinical Course in Patients with IgG4-Related Ophthalmic Disease by Unbiased Clustering Analysis. J Clin Med 9:4084. https://doi.org/10.3390/jcm9124084
Varghese JS, Ho JC, Anjana RM et al (2021) Profiles of Intraday Glucose in Type 2 Diabetes and Their Association with Complications: An Analysis of Continuous Glucose Monitoring Data. Diabetes Technol Ther 23:555–564. https://doi.org/10.1089/dia.2020.0672
Velez G, Roybal CN, Colgan D et al (2016) Precision Medicine: Personalized Proteomics for the Diagnosis and Treatment of Idiopathic Inflammatory Disease. JAMA Ophthalmol 134:444–448. https://doi.org/10.1001/jamaophthalmol.2015.5934
Velez G, Roybal CN, Binkley E et al (2017) Proteomic analysis of elevated intraocular pressure with retinal detachment. Am J Ophthalmol Case Rep 5:107–110. https://doi.org/10.1016/j.ajoc.2016.12.023
Velez G, Yang J, Li AS et al (2019) Proteomic insight into the pathogenesis of CAPN5-vitreoretinopathy. Sci Rep 9:7608. https://doi.org/10.1038/s41598-019-44031-7
Wang H, Zhai R, Sun Q et al (2019) Metabolomic Profile of Posner-Schlossman Syndrome: A Gas Chromatography Time-of-Flight Mass Spectrometry-Based Approach Using Aqueous Humor. Front Pharmacol 10:1–11. https://doi.org/10.3389/fphar.2019.01322
Wang H, Fang J, Chen F et al (2020) Metabolomic profile of diabetic retinopathy: a GC-TOFMS-based approach using vitreous and aqueous humor. Acta Diabetol 57:41–51. https://doi.org/10.1007/s00592-019-01363-0
Wang W, Pei X, Zhang L et al (2021) Application of new international classification of adult-onset diabetes in Chinese inpatients with diabetes mellitus. Diabetes Metab Res Rev 37:1–8. https://doi.org/10.1002/dmrr.3427
Wei Q, Jiang C, Ye X et al (2019) Vitreous Proteomics Provides New Insights into Antivascular Endothelial Growth Factor Therapy for Pathologic Myopia Choroid Neovascularization. J Interf Cytokine Res 39:786–796. https://doi.org/10.1089/jir.2019.0030
Wei P, He M, Teng H, Han G (2020) Metabolomic analysis of the aqueous humor from patients with central retinal vein occlusion using UHPLC-MS/MS. J Pharm Biomed Anal 188:113448. https://doi.org/10.1016/j.jpba.2020.113448
Wen K, Shao X, Li Y et al (2021) The plasminogen protein is associated with high myopia as revealed by the iTRAQ-based proteomic analysis of the aqueous humor. Sci Rep 11:8789. https://doi.org/10.1038/s41598-021-88220-9
Wu Z, Ding N, Yu M et al (2016) Identification of Potential Biomarkers for Rhegmatogenous Retinal Detachment Associated with Choroidal Detachment by Vitreous iTRAQ-Based Proteomic Profiling. Int J Mol Sci 17:2052. https://doi.org/10.3390/ijms17122052
Wu X, Liu Z, Zhang X et al (2017) Proteomics analysis and proteogenomic characterization of different physiopathological human lenses. BMC Ophthalmol 17:253. https://doi.org/10.1186/s12886-017-0642-9
Xiao H, Xin W, Sun LM et al (2021) Comprehensive proteomic profiling of aqueous humor proteins in proliferative diabetic retinopathy. Transl Vis Sci Technol 10:0–7. https://doi.org/10.1167/tvst.10.6.3
Xiong X, Yang Y, Wei L et al (2021) Identification of two novel subgroups in patients with diabetes mellitus and their association with clinical outcomes: A two-step cluster analysis. J Diabetes Investig 12:1346–1358. https://doi.org/10.1111/jdi.13494
Xuan Q, Zheng F, Yu D et al (2020) Rapid lipidomic profiling based on ultra-high performance liquid chromatography–mass spectrometry and its application in diabetic retinopathy. Anal Bioanal Chem 412:3585–3594. https://doi.org/10.1007/s00216-020-02632-6
Yam GH-F, Fuest M, Zhou L et al (2019) Differential epithelial and stromal protein profiles in cone and non-cone regions of keratoconus corneas. Sci Rep 9:2965. https://doi.org/10.1038/s41598-019-39182-6
Yamamoto Y, Morozumi T, Hirata T et al (2020) Effect of Periodontal Disease on Diabetic Retinopathy in Type 2 Diabetic Patients: A Cross-Sectional Pilot Study. J Clin Med 9:3234. https://doi.org/10.3390/jcm9103234
Yang H, Yang X, Wang Y et al (2020) Comparative analysis of the tear protein profile in herpes simplex virus type 1 epithelial keratitis. BMC Ophthalmol 20:355. https://doi.org/10.1186/s12886-020-01626-3
Yao J, Chen Z, Yang Q et al (2013) Proteomic analysis of aqueous humor from patients with branch retinal vein occlusion-induced macular edema. Int J Mol Med 32:1421–1434. https://doi.org/10.3892/ijmm.2013.1509
Yao J, Liu X, Yang Q et al (2013) Proteomic analysis of the aqueous humor in patients with wet age-related macular degeneration. PROTEOMICS - Clin Appl 7:550–560. https://doi.org/10.1002/prca.201200012
Yawata N, Awate S, Liu Y-C et al (2019) Kinetics of Tear Fluid Proteins after Endothelial Keratoplasty and Predictive Factors for Recovery from Corneal Haze. J Clin Med 9:63. https://doi.org/10.3390/jcm9010063
Young SP, Nessim M, Falciani F et al (2009) Metabolomic analysis of human vitreous humor differentiates ocular inflammatory disease. Mol Vis 15:1210–1217
Yu J, Feng L, Wu Y et al (2014) Vitreous proteomic analysis of idiopathic epiretinal membranes. Mol BioSyst 10:2558–2566. https://doi.org/10.1039/C4MB00240G
Yu M, Wu Z, Zhang Z et al (2015) Metabolomic Analysis of Human Vitreous in Rhegmatogenous Retinal Detachment Associated With Choroidal Detachment. Investig Opthalmology Vis Sci 56:5706. https://doi.org/10.1167/iovs.14-16338
Zhang L, Masetti G, Colucci G et al (2018) Combining micro-RNA and protein sequencing to detect robust biomarkers for Graves’ disease and orbitopathy. Sci Rep 8:8386. https://doi.org/10.1038/s41598-018-26700-1
Zhang D, Zhao Y, Yang Y-T et al (2020) A Mechanism Study of Electroacupuncture for Dry Eye Syndrome by Targeting Conjunctival Cytokine Expressions. Curr Eye Res 45:419–427. https://doi.org/10.1080/02713683.2019.1666997
Zhang S, Zhang K, He W et al (2021) Quantitative Phosphoproteomic Comparison of Lens Proteins in Highly Myopic Cataract and Age-Related Cataract. Biomed Res Int 2021:1–18. https://doi.org/10.1155/2021/6668845
Zhavoronkov A, Kanherkar RR, Izumchenko E et al (2016) Pro-fibrotic pathway activation in trabecular meshwork and lamina cribrosa is the main driving force of glaucoma. Cell Cycle 15:1643–1652. https://doi.org/10.1080/15384101.2016.1170261
Zhou L, Zhao SZ, Koh SK et al (2012) In-depth analysis of the human tear proteome. J Proteomics 75:3877–3885. https://doi.org/10.1016/j.jprot.2012.04.053
Zhu X, Du Y, Truscott RJW et al (2020) Profiling and Bioinformatic Analysis of Differentially Expressed Cytokines in Aqueous Humor of High Myopic Eyes – Clues for Anti-VEGF Injections. Curr Eye Res 45:97–103. https://doi.org/10.1080/02713683.2019.1648833
Zou C, Zhao M, Yu J et al (2018) Difference in the Vitreal Protein Profiles of Patients with Proliferative Diabetic Retinopathy with and without Intravitreal Conbercept Injection. J Ophthalmol 2018:1–11. https://doi.org/10.1155/2018/7397610
Zou X, Wang S, Zhang P et al (2020) Quantitative Proteomics and Weighted Correlation Network Analysis of Tear Samples in Adults and Children With Diabetes and Dry Eye. Transl Vis Sci Technol 9:8. https://doi.org/10.1167/tvst.9.13.8
Zuo J, Lan Y, Hu H et al (2021) Metabolomics-based multidimensional network biomarkers for diabetic retinopathy identification in patients with type 2 diabetes mellitus. BMJ Open Diabetes Res Care 9:e001443. https://doi.org/10.1136/bmjdrc-2020-001443
Funding
This study was in part supported by the Fighting Blindness Canada’s (FBC’s) Clinician Scientist Emerging Leader Award granted to Dr. Tina Felfeli.
Author information
Authors and Affiliations
Contributions
Conceptualization: Saffire H Krance, Karthik Manichavagan, Rafael N Miranda, Tina Felfeli; Methodology: Saffire H Krance, Aidan Pucchio, Karthik Manichavagan, Rafael N Miranda, Tina Felfeli; Formal analysis and investigation: Saffire H Krance, Aidan Pucchio, Daiana R Pur, Arshpreet Bassi, Jasmine Bhatti, Karthik Manichavagan, Shaily Brahmbhatt, Ishita Aggarwal, Priyanka Singh, Aleena Virani, Meagan Stanley, Rafael N Miranda, Tina Felfeli; Writing—original draft preparation: Saffire H Krance, Aidan Pucchio, Daiana R Pur, Arshpreet Bassi, Rafael N Miranda, Tina Felfeli; Writing—review and editing: Saffire H Krance, Aidan Pucchio, Rafael N Miranda, Tina Felfeli; Funding acquisition: Tina Felfeli; Supervision: Rafael N Miranda, Tina Felfeli.
Corresponding author
Ethics declarations
Ethics approval
As this is a scoping review, no human participants were involved, and IRB approval was not required.
Conflict of interest
No conflicting relationship exists for any author. The authors do not have any proprietary interests in the materials described in the article.
Presentations
The contents of this manuscript have not been presented at any conferences priorto submission.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Aidan Pucchio and Saffire H Krance are co-first authors.
Appendices
Appendix A
Search strategy utilized for five electronic databases (EMBASE, Medline, Cochrane Central Register of Controlled Trials, Cochrane Database of Systematic Reviews, Web of Science).
Embase
-
1.
(ophth* or ocular or intraocular or eye* or retina* or macula* or fovea* or uvea* or sclera* or cornea* or conjunctiva* or iris or "vitreous body" or "vitreous humo?r" or "vitreous fluid" or vitreo* or "aqueous humo?r" or "aqueous fluid" or tears or ((tear or lacrimal) adj fluid) or glaucoma or retinop* or retinoblastoma or uveitis or iritis or choroiditis or retinitis or chorioretinitis or conjunctivitis or endophthalmitis or cataract* or ?????????opia or "optic atrophy" or "optic neuropathy" or vitrectomy or phacoemulsification or trabeculotomy or (paracentesis adj3 "anterior chamber")).tw.
-
2.
("precision medicine" or "precision health" or "personalized medicine" or "personalized proteomics" or thera?nostic? or "tailored medicine" or "artificial intelligence" or "machine learning" or "deep learning" or algorithm? or ((supervised or unsupervised or biased or unbiased or bayesian or hierarchical or neur??al) adj (cluster* or learning or learner? or classifi* or network?)) or "k-nearest neighbo?r?" or "naive bayes" or (decision adj (tree? or forest? or jungle?)) or "random forest?" or "gradient-boost*" or "support vector machine" or "k-means" or "association rules" or "recursive partitioning" or "discriminant analysis" or "feature selection" or ((linear or nonlinear or "non-linear" or logistic or ordinal or poisson or quantile or analysis) adj1 (regression? or model?)) or bioinformatic? or "gene ontology" or "Kyoto Encyclopedia of Genes and Genomes" or "KEGG" or ((progress* or regress* or recover* or respond* or response*) and (predict* or stratif*))).tw.
-
3.
(Proteomic? or proteome? or metabolomic? or metabolome? or lipidomic? or lipidome? or "?????inflammatory protein?" or "?????inflammatory marker" or cytokine? or interleukin? or lymphokine? or monokine? or interferon? or "colony stimulating factor?" or chemokine? or "growth factor?" or "necrosis factor?" or "chemotactic protein?" or "adhesion molecule?" or "adhesion protein?" or "matrix metalloproteinase-2" or myeloperoxidase? or "tissue inhibitor of metalloproteinase-2" or "macrophage inflammatory protein-1" or "brain-derived neurotrophic factor" or angiopoietin? or ((hemoglobin or haemoglobin) adj1 (a1c or glycated)) or hba1c or "c reactive protein" or "c-reactive protein" or crp or hscrp or "hs-crp" or ((protein or biomarker) and (concentration? or level? or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay))).tw.
-
4.
ophthalmology/
-
5.
eye/ or anterior eye chamber/ or anterior eye segment/ or aqueous humor/ or exp conjunctiva/ or exp cornea/ or eye fundus/ or eyeball/ or exp lens/ or ocular blood vessel/ or ophthalmic artery/ or optic disk/ or palpebral fissure/ or posterior eye chamber/ or posterior eye segment/ or exp retina/ or exp sclera/ or sphincter pupillae muscle/ or tenon capsule/ or trabecular meshwork/ or exp uvea/ or vitreous body/
-
6.
lacrimal fluid/
-
7.
eye disease/ or exp accommodation disorder/ or exp conjunctiva disease/ or exp cornea disease/ or exp dry eye/ or exp eye burning/ or exp eye chamber disease/ or exp eye discharge/ or exp eye discomfort/ or exp eye edema/ or exp eye infection/ or exp eye inflammation/ or exp eye injury/ or exp eye irritation/ or exp eye jaundice/ or exp eye malformation/ or exp eye pain/ or exp eye redness/ or exp eye swelling/ or exp eye toxicity/ or exp eye tumor/ or exp glaucoma/ or exp intraocular hemorrhage/ or exp intraocular pressure abnormality/ or exp lens disease/ or exp ocular albinism/ or exp ocular fibrosis/ or exp ocular pruritus/ or exp ocular surface disease/ or exp optic nerve disease/ or exp photophobia/ or exp pupil disease/ or exp retina disease/ or exp sclera disease/ or exp uvea disease/ or exp visual disorder/ or exp vitreous disease/
-
8.
exp vitrectomy/
-
9.
exp phacoemulsification/
-
10.
exp trabeculectomy/
-
11.
personalized medicine/
-
12.
theranostic nanomedicine/
-
13.
algorithm/
-
14.
exp clustering algorithm/
-
15.
artificial intelligence/
-
16.
exp machine learning/
-
17.
"decision tree"/
-
18.
bioinformatics/
-
19.
exp regression analysis/
-
20.
discriminant analysis/
-
21.
gene ontology/
-
22.
proteomics/ or comparative proteomics/ or immunoproteomics/ or exp pharmacoproteomics/ or phosphoproteomics/ or proteogenomics/ or secretomics/
-
23.
proteome/
-
24.
metabolomics/
-
25.
metabolome/
-
26.
lipidomics/
-
27.
lipidome/
-
28.
exp cytokine/
-
29.
exp cell adhesion molecule/
-
30.
myeloperoxidase/
-
31.
"tissue inhibitor of metalloproteinase 1"/
-
32.
"tissue inhibitor of metalloproteinase 2"/
-
33.
brain derived neurotrophic factor/
-
34.
exp angiopoietin/
-
35.
exp "peptides and proteins"/ec [Endogenous Compound]
-
36.
biological marker/ec [Endogenous Compound]
-
37.
1 or 4 or 5 or 6 or 7 or 8 or 9 or 10
-
38.
2 or 11 or 12 or 13 or 14 or 15 or 16 or 17 or 18 or 19 or 20 or 21
-
39.
(concentration? or level? or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay).tw.
-
40.
(35 or 36) and 39
-
41.
3 or 22 or 23 or 24 or 25 or 26 or 27 or 28 or 29 or 30 or 31 or 32 or 33 or 34 or 40
-
42.
37 and 38 and 41
-
43.
limit 42 to conference abstracts
-
44.
limit 42 to animal studies
-
45.
limit 44 to human
-
46.
limit 42 to "review"
-
47.
44 not 45
-
48.
42 not (43 or 46 or 47)
Medline
-
1.
(ophth* or ocular or intraocular or eye* or retina* or macula* or fovea* or uvea* or sclera* or cornea* or conjunctiva* or iris or "vitreous body" or "vitreous humo?r" or "vitreous fluid" or vitreo* or "aqueous humo?r" or "aqueous fluid" or tears or ((tear or lacrimal) adj fluid) or glaucoma or retinop* or retinoblastoma or uveitis or iritis or choroiditis or retinitis or chorioretinitis or conjunctivitis or endophthalmitis or cataract* or ?????????opia or "optic atrophy" or "optic neuropathy" or vitrectomy or phacoemulsification or trabeculotomy or (paracentesis adj3 "anterior chamber")).tw.
-
2.
("precision medicine" or "precision health" or "personalized medicine" or "personalized proteomics" or thera?nostic? or "tailored medicine" or "artificial intelligence" or "machine learning" or "deep learning" or algorithm? or ((supervised or unsupervised or biased or unbiased or bayesian or hierarchical or neur??al) adj (cluster* or learning or learner? or classifi* or network?)) or "k-nearest neighbo?r?" or "naive bayes" or (decision adj (tree? or forest? or jungle?)) or "random forest?" or "gradient-boost*" or "support vector machine" or "k-means" or "association rules" or "recursive partitioning" or "discriminant analysis" or "feature selection" or ((linear or nonlinear or "non-linear" or logistic or ordinal or poisson or quantile or analysis) adj1 (regression? or model?)) or bioinformatic? or "gene ontology" or "Kyoto Encyclopedia of Genes and Genomes" or "KEGG" or ((progress* or regress* or recover* or respond* or response*) and (predict* or stratif*))).tw.
-
3.
(Proteomic? or proteome? or metabolomic? or metabolome? or lipidomic? or lipidome? or "?????inflammatory protein?" or "?????inflammatory marker" or cytokine? or interleukin? or lymphokine? or monokine? or interferon? or "colony stimulating factor?" or chemokine? or "growth factor?" or "necrosis factor?" or "chemotactic protein?" or "adhesion molecule?" or "adhesion protein?" or "matrix metalloproteinase-2" or myeloperoxidase? or "tissue inhibitor of metalloproteinase-2" or "macrophage inflammatory protein-1" or "brain-derived neurotrophic factor" or angiopoietin? or ((hemoglobin or haemoglobin) adj1 (a1c or glycated)) or hba1c or "c reactive protein" or "c-reactive protein" or crp or hscrp or "hs-crp" or ((protein or biomarker) and (concentration? or level? or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay))).tw.
-
4.
exp Ophthalmology/cl, di, dg, ec, pd, px, sn, sd, su, th, td, ed, es, hi, is, mt, og, rt, st [Classification, Diagnosis, Diagnostic Imaging, Economics, Pharmacology, Psychology, Statistics & Numerical Data, Supply & Distribution, Surgery, Therapy, Trends, Education, Ethics, History, Instrumentation, Methods, Organization & Administration, Radiotherapy, Standards]
-
5.
eye/ or exp anterior eye segment/ or "anterior capsule of the lens"/ or conjunctiva/ or meibomian glands/ or exp "pigment epithelium of eye"/ or exp posterior eye segment/ or exp retina/ or sclera/ or tenon capsule/ or exp uvea/
-
6.
Tears/
-
7.
eye diseases/ or cogan syndrome/ or exp conjunctival diseases/ or exp corneal diseases/ or exp eye abnormalities/ or exp eye diseases, hereditary/ or exp eye hemorrhage/ or exp eye infections/ or exp eye injuries/ or exp eye manifestations/ or exp eye neoplasms/ or exp lens diseases/ or exp ocular hypertension/ or ocular hypotension/ or exp optic nerve diseases/ or exp pupil disorders/ or exp refractive errors/ or exp retinal diseases/ or exp scleral diseases/ or exp uveal diseases/ or exp vision disorders/ or vitreous detachment/
-
8.
Vitrectomy/ae, ec, ed, es, hi, is, mt, mo, nu, px, rh, st, sn, td [Adverse Effects, Economics, Education, Ethics, History, Instrumentation, Methods, Mortality, Nursing, Psychology, Rehabilitation, Standards, Statistics & Numerical Data, Trends]
-
9.
Phacoemulsification/ae, cl, ec, ed, hi, is, mt, mo, nu, px, rh, st, sn, td [Adverse Effects, Classification, Economics, Education, History, Instrumentation, Methods, Mortality, Nursing, Psychology, Rehabilitation, Standards, Statistics & Numerical Data, Trends]
-
10.
Trabeculectomy/nu, px, rh, st, sn, td, ae, cl, ec, ed, hi, is, mt, mo [Nursing, Psychology, Rehabilitation, Standards, Statistics & Numerical Data, Trends, Adverse Effects, Classification, Economics, Education, History, Instrumentation, Methods, Mortality]
-
11.
Precision Medicine/ae, cl, ec, es, hi, is, mt, mo, nu, px, st, sn, td [Adverse Effects, Classification, Economics, Ethics, History, Instrumentation, Methods, Mortality, Nursing, Psychology, Standards, Statistics & Numerical Data, Trends]
-
12.
Theranostic Nanomedicine/
-
13.
exp algorithms/
-
14.
Neural Networks, Computer/
-
15.
Decision Trees/
-
16.
exp Regression Analysis/
-
17.
Discriminant Analysis/
-
18.
exp Proteomics/cl, ec, ed, es, hi, is, mt, og, st, sn, td [Classification, Economics, Education, Ethics, History, Instrumentation, Methods, Organization & Administration, Standards, Statistics & Numerical Data, Trends]
-
19.
Proteome/
-
20.
exp Metabolomics/cl, ec, ed, es, hi, is, mt, og, st, sn, td [Classification, Economics, Education, Ethics, History, Instrumentation, Methods, Organization & Administration, Standards, Statistics & Numerical Data, Trends]
-
21.
Metabolome/
-
22.
exp Cytokines/
-
23.
exp Cell Adhesion Molecules/
-
24.
Matrix Metalloproteinase 2/
-
25.
Peroxidase/
-
26.
"Tissue Inhibitor of Metalloproteinase-1"/
-
27.
"Tissue Inhibitor of Metalloproteinase-2"/Brain-Derived Neurotrophic Factor/
-
28.
exp Angiopoietins/
-
29.
Gene Ontology/
-
30.
exp Proteins/
-
31.
exp Peptides/
-
32.
Biomarkers/
-
33.
(concentration? or level? or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay).tw.
-
34.
(31 or 32 or 33) and 3
-
35.
1 or 4 or 5 or 6 or 7 or 8 or 9 or 10
-
36.
2 or 11 or 12 or 13 or 14 or 15 or 16 or 17 or 30
-
37.
3 or 18 or 19 or 20 or 21 or 22 or 23 or 24 or 25 or 26 or 27 or 28 or 29 or 35
-
38.
36 and 37 and 38
-
39.
limit 39 to animals
-
40.
limit 40 to humans
-
41.
40 not 41
-
42.
limit 39 to "review articles"
-
43.
39 not (42 or 43)
Web of science
-
1.
(TI = (ophth* or ocular or intraocular or eye* or retina* or macula* or fovea* or uvea* or sclera* or cornea* or conjunctiva* or iris or "vitreous body" or "vitreous humo$r" or "vitreous fluid" or vitreo* or "aqueous humo$r" or "aqueous fluid" or tears or ((tear or lacrimal) NEAR/1 fluid) or glaucoma or retinop* or retinoblastoma or uveitis or iritis or choroiditis or retinitis or chorioretinitis or conjunctivitis or endophthalmitis or cataract* or *opia or "optic atrophy" or "optic neuropathy" or vitrectomy or phacoemulsification or trabeculotomy.tw. or (paracentesis NEAR/3 "anterior chamber")) or AB = (ophth* or ocular or intraocular or eye* or retina* or macula* or fovea* or uvea* or sclera* or cornea* or conjunctiva* or iris.tw. or "vitreous body" or "vitreous humo$r" or "vitreous fluid" or vitreo* or "aqueous humo$r" or "aqueous fluid" or tears or ((tear or lacrimal) NEAR/1 fluid) or glaucoma or retinop* or retinoblastoma or uveitis or iritis or choroiditis or retinitis or chorioretinitis or conjunctivitis or endophthalmitis or cataract* or "optic atrophy" or "optic neuropathy" or vitrectomy or phacoemulsification or trabeculotomy or (paracentesis NEAR/3 "anterior chamber"))) AND
-
2.
(TI = ("precision medicine" or "precision health" or "personalized medicine" or "personalized proteomics" or thera$nostic* or "tailored medicine" or "artificial intelligence" or "machine learning" or "deep learning" or algorithm$ or ((supervised or unsupervised or biased or unbiased or bayesian or hierarchical or neural or neuronal) NEAR/1 (cluster* or learning or learner$ or classifi* or network$)) or "k-nearest neighbo$r*" or "naive bayes" or (decision NEAR/1 (tree$ or forest$ or jungle$)) or "random forest$" or "gradient-boost*" or "support vector machine" or "k-means" or "association rules" or "recursive partitioning" or "discriminant analysis" or "feature selection" or ((linear or nonlinear or "non-linear" or logistic or ordinal or poisson or quantile or analysis) NEAR/1 (regression$ or model$)) or bioinformatic$ or ((progress* or regress* or recover* or respond* or response*) and (predict* or stratif*))) OR AB = ("precision medicine" or "precision health" or "personalized medicine" or "personalized proteomics" or thera$nostic* or "tailored medicine" or "artificial intelligence" or "machine learning" or "deep learning" or algorithm$ or ((supervised or unsupervised or biased or unbiased or bayesian or hierarchical or neural or neuronal) NEAR/1 (cluster* or learning or learner$ or classifi* or network$)) or "k-nearest neighbo$r*" or "naive bayes" or (decision NEAR/1 (tree$ or forest$ or jungle$)) or "random forest$" or "gradient-boost*" or "support vector machine" or "k-means" or "association rules" or "recursive partitioning" or "discriminant analysis" or "feature selection" or ((linear or nonlinear or "non-linear" or logistic or ordinal or poisson or quantile or analysis) NEAR/1 (regression$ or model$)) or bioinformatic$ or ((progress* or regress* or recover* or respond* or response*) and (predict* or stratif*)))) AND
-
3.
(TI = (Proteomic$ or proteome$ or metabolomic$ or metabolome$ or lipidomic$ or lipidome$ or "*inflammatory protein$" or "*inflammatory marker" or cytokine$ or interleukin$ or lymphokine$ or monokine$ or interferon$ or "colony stimulating factor$" or chemokine$ or "growth factor$" or "necrosis factor$" or "chemotactic protein$" or "adhesion molecule$" or "adhesion protein$" or "matrix metalloproteinase-2" or myeloperoxidase$ or "tissue inhibitor of metalloproteinase-2" or "macrophage inflammatory protein-1" or "brain-derived neurotrophic factor" or angiopoietin$ or ((hemoglobin or haemoglobin) NEAR/1 (a1c or glycated or glycosylated)) or hba1c or "c reactive protein" or "c-reactive protein" or crp or hscrp or "hs-crp" or ((protein or biomarker) and (concentration$ or level$ or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay))) OR AB = (Proteomic$ or proteome$ or metabolomic$ or metabolome$ or lipidomic$ or lipidome$ or "inflammatory protein$" or "inflammatory marker" or cytokine$ or interleukin$ or lymphokine$ or monokine$ or interferon$ or "colony stimulating factor$" or chemokine$ or "growth factor$" or "necrosis factor$" or "chemotactic protein$" or "adhesion molecule$" or "adhesion protein$" or "matrix metalloproteinase-2" or myeloperoxidase$ or "tissue inhibitor of metalloproteinase-2" or "macrophage inflammatory protein-1" or "brain-derived neurotrophic factor" or angiopoietin$ or ((hemoglobin or haemoglobin) NEAR/1 (a1c or glycated or glycosylated)) or hba1c or "c reactive protein" or "c-reactive protein" or crp or hscrp or "hs-crp" or ((protein or biomarker) and (concentration$ or level$ or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay))))
Cochrane central register of controlled trials (CONTROL), cochrane database of systematic reviews
-
1.
ophth* or ocular or intraocular or eye* or retina* or macula* or fovea* or uvea* or sclera* or cornea* or conjunctiva* or iris or "vitreous body" or "vitreous humo*r" or "vitreous fluid" or vitreo* or "aqueous humo*r" or "aqueous fluid" or tears or ((tear or lacrimal) NEXT fluid) or glaucoma or retinop* or retinoblastoma or uveitis or iritis or choroiditis or retinitis or chorioretinitis or conjunctivitis or endophthalmitis or cataract* or *opia or "optic atrophy" or "optic neuropathy" or vitrectomy or phacoemulsification or trabeculotomy or (paracentesis NEAR/3 "anterior chamber")
-
2.
"precision medicine" or "precision health" or "personalized medicine" or "personalized proteomics" or thera*nostic* or "tailored medicine" or "artificial intelligence" or "machine learning" or "deep learning" or algorithm* or ((supervised or unsupervised or biased or unbiased or bayesian or hierarchical or neural or neuronal) NEXT (cluster* or learning or learner* or classifi* or network*)) or "k-nearest neighbo*r*" or "naive bayes" or (decision NEXT (tree* or forest* or jungle*)) or "random forest*" or "gradient-boost*" or "support vector machine" or "k-means" or "association rules" or "recursive partitioning" or "discriminant analysis" or "feature selection" or ((linear or nonlinear or "non-linear" or logistic or ordinal or poisson or quantile or analysis) NEXT (regression* or model*)) or bioinformatic* or ((progress* or regress* or recover* or respond* or response*) and (predict* or stratif*))
-
3.
Proteomic* or proteome* or metabolomic* or metabolome* or lipidomic* or lipidome* or "*inflammatory protein*" or "*inflammatory marker*" or cytokine* or interleukin* or lymphokine* or monokine$ or interferon* or "colony stimulating factor*" or chemokine* or "growth factor*" or "necrosis factor*" or "chemotactic protein*" or "adhesion molecule*" or "adhesion protein*" or "matrix metalloproteinase-2" or myeloperoxidase* or "tissue inhibitor of metalloproteinase-2" or "macrophage inflammatory protein-1" or "brain-derived neurotrophic factor" or angiopoietin* or ((hemoglobin or haemoglobin) NEXT (a1c or glycated or glycosylated)) or hba1c or "c reactive protein" or "c-reactive protein" or crp or hscrp or "hs-crp" or ((protein or biomarker) and (concentration* or level* or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay))
-
4.
MeSH descriptor: [Ophthalmology]
-
5.
MeSH descriptor: [Eye]
-
6.
MeSH descriptor: [Tears]
-
7.
MeSH descriptor: [Eye Diseases]
-
8.
MeSH descriptor: [Vitrectomy]
-
9.
MeSH descriptor: [Phacoemulsification]
-
10.
MeSH descriptor: [Trabeculectomy]
-
11.
MeSH descriptor: [Precision Medicine]
-
12.
MeSH descriptor: [Theranostic Nanomedicine]
-
13.
MeSH descriptor: [Artificial Intelligence]
-
14.
MeSH descriptor: [Algorithms]
-
15.
MeSH descriptor: [Neural Networks, Computer]
-
16.
MeSH descriptor: [Decision Trees]
-
17.
MeSH descriptor: [Regression Analysis]
-
18.
MeSH descriptor: [Discriminant Analysis]
-
19.
MeSH descriptor: [Proteomics]
-
20.
MeSH descriptor: [Metabolomics]
-
21.
MeSH descriptor: [Proteins] explode
-
22.
MeSH descriptor: [Peptides]
-
23.
MeSH descriptor: [Biomarkers]
-
24.
concentration* or level* or quantif* or quantit* or mass spectrometry or iTRAQ or MALDI or SELDI or assay
-
25.
(#21 or #22 or #23) and #24
-
26.
MeSH descriptor: [Cytokines]
-
27.
MeSH descriptor: [Cytokines]
-
28.
MeSH descriptor: [Matrix Metalloproteinase 2]
-
29.
MeSH descriptor: [Peroxidase]
-
30.
MeSH descriptor: [Tissue Inhibitor of Metalloproteinase-1]
-
31.
MeSH descriptor: [Tissue Inhibitor of Metalloproteinase-2]
-
32.
MeSH descriptor: [Brain-Derived Neurotrophic Factor
-
33.
MeSH descriptor: [Angiopoietins]
-
34.
MeSH descriptor: [Anterior Eye Segment]
-
35.
MeSH descriptor: [Anterior Capsule of the Lens]
-
36.
MeSH descriptor: [Axial Length, Eye]
-
37.
MeSH descriptor: [Pigment Epithelium of Eye]
-
38.
MeSH descriptor: [Posterior Eye Segment]
-
39.
MeSH descriptor: [Retina]
-
40.
MeSH descriptor: [Sclera]
-
41.
MeSH descriptor: [Tenon Capsule]
-
42.
MeSH descriptor: [Uvea]
-
43.
MeSH descriptor: [Asthenopia]
-
44.
MeSH descriptor: [Cogan Syndrome]
-
45.
MeSH descriptor: [Conjunctival Diseases]
-
46.
MeSH descriptor: [Corneal Diseases]
-
47.
MeSH descriptor: [Eye Abnormalities]
-
48.
MeSH descriptor: [Eye Diseases, Hereditary]
-
49.
MeSH descriptor: [Eye Hemorrhage]
-
50.
MeSH descriptor: [Eye Infections]
-
51.
MeSH descriptor: [Eye Injuries]
-
52.
MeSH descriptor: [Eye Manifestations]
-
53.
MeSH descriptor: [Eye Neoplasms]
-
54.
MeSH descriptor: [Lens Diseases]
-
55.
MeSH descriptor: [Ocular Hypertension]
-
56.
MeSH descriptor: [Ocular Hypotension]
-
57.
MeSH descriptor: [Optic Nerve Diseases]
-
58.
MeSH descriptor: [Pupil Disorders]
-
59.
MeSH descriptor: [Refractive Errors]
-
60.
MeSH descriptor: [Retinal Diseases]
-
61.
MeSH descriptor: [Scleral Diseases]
-
62.
MeSH descriptor: [Uveal Diseases]
-
63.
MeSH descriptor: [Vision Disorders]
-
64.
MeSH descriptor: [Vitreous Detachment]
-
65.
#1 or #4 or #5 or #6 or #7 or #8 or #9 or #10 or #34 or #35 or #36 or #37 or #38 or #39 or #40 or #41 or #42 or #43 or #44 or #45 or #46 or #47 or #48 or #49 or #50 or #51 or #52 or #53 or #54 or #55 or #56 or #57 or #58 or #59 or #60 or #61 or #62 or #63 or #64
-
66.
#2 or #11 or #12 or #13 or #14 or #15 or #16 or #17 or #18
-
67.
#3 or #19 or #20 or #25 or #26 or #27 or #28 or #29 or #30 or #31 or #32 or #33
-
68.
#65 and #66 and #67
Appendix B
Table
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pucchio, A., Krance, S.H., Pur, D.R. et al. Applications of artificial intelligence and bioinformatics methodologies in the analysis of ocular biofluid markers: a scoping review. Graefes Arch Clin Exp Ophthalmol 262, 1041–1091 (2024). https://doi.org/10.1007/s00417-023-06100-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-023-06100-6