Abstract
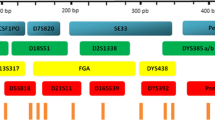
The HomyGene19+14Y System (HG19+14Y) is a PCR-based amplification kit that enables typing of 18 autosomal short tandem repeat (STR) loci (i.e., CSF1PO, D2S1338, D3S1358, D5S818, D6S1043, D7S820, D8S1179, D12S391, D13S317, D16S539, D18S51, D19S433, D21S11, FGA, Penta E, TPOX, TH01, vWA), 14 widely used Y chromosome STR (Y-STR) loci (Y_GATA_H4, DYS385a/b, DYS389I/II, DYS390, DYS391, DYS392, DYS393, DYS438, DYS439, DYS456, DYS458, DYS635), and amelogenin. This multiplex system was designed for the simultaneous analysis of amelogenin-Y allele mutation, single-source searches, kinship (including familial searching), mixture profiles, international data sharing, and other forensic applications. In this study, the multiplex system was validated for sensitivity, specificity, DNA mixtures, stability, precision, stutter, reproducibility, parallel tests, PCR-based conditions, and population analysis according to the Scientific Working Group on DNA Analysis Methods (SWGDAM) developmental validation guidelines. A total of 212 alleles were detected for the 18 autosomal STR loci among 528 Guangdong Han individuals, and 431 haplotypes were found for 14 Y-STRs among 452 unrelated males. The combined match probability (CMP) of the HG19+14Y System was calculated as 2.39 × 10−29. All the validation results showed that the HG19+14Y System would be a robust, reliable, highly polymorphic, and informative forensic kit.












Similar content being viewed by others
References
Budowle B (1997) Studies for selecting core STR loci for CODIS in presented at DNA forensics science, evidence, and future prospects. November 17–18, McLean, VA
Butler JM (2006) Genetics and genomics of core short tandem repeat loci used in human identity testing. J Forensic Sci 51:253–265
Jiang XH (2011) Role of technologies in national DNA database in China: past, present and future trends. Chin. J Forensic Med 26:383–386
Ensenberger MG, Lenz KA, Matthies LK, Hadinoto GM et al (2016) Developmental validation of the PowerPlex Fusion 6C System. Forensic Sci Int Genet 21:134–144
Wang DY, Gopinath S, Lagace RE, Norona W, Hennessy LK, Short ML, Mulero JJ (2015) Developmental validation of the GlobalFiler Express PCR Amplification Kit: a 6-dye multiplex assay for the direct amplification of reference samples. Forensic Sci Int Genet 19:148–155
Thompson JM, Ewing MM, Frank WE, Pogemiller JJ et al (2013) Developmental validation of the PowerPlex Y23 System: a single multiplex Y-STR analysis system for casework and database samples. Forensic Sci Int Genet 7:240–250
Gopinath S, Zhong C, Nguyen V, Ge JY, Lagace RE, Short ML, Mulero JJ (2016) Developmental validation of the Yfiler Plus PCR Ampli fication Kit: an enhanced Y-STR multiplex for casework and database applications. Forensic Sci Int Genet 24:164–175
Hares DR (2012) Expanding the CODIS core loci in the United States. Forensic Sci Int Genet 6(1):52–54
Ge JY, Eisenberg A, Budowle B (2012) Developing criteria and data to determine best options for expanding the core CODIS loci. Invest Genet 3:1–48
Gill P, Haned H, Bleka O, Hansson O, Dorum G, Egeland T (2015) Genotyping and interpretation of STR-DNA: low-template, mixtures and database matches—twenty years of research and development. Forensic Sci Int Genet 18:100–117
Butler JM, Shen Y, McCord BR (2003) The development of reduced size STR amplicons as tools for analysis of degraded DNA. J Forensic Sci 48(5):1054–1064
Ballantyne KN, Oorschot RA, Mitchell RJ (2007) Comparison of two whole genome amplification methods for STR genotyping of LCN and degraded DNA samples. Forensic Sci Int 166:35–41
Lopes V, Andrade L, Carvalho M, Serra A, Balsa F, Bento AM et al (2009) Mini-STRs a powerful tool to identify genetic profiles in samples with small amounts of DNA. Forensic Sci Int Genet 2:121–122
Guo F, Zhou YS, Jiang XH et al (2016) Evaluation of the Early Access STR Kit v1 on the Ion Torrent PGM™ platform. Forensic Sci Int Genet 23:111–120
Martin PD, Schmitter H, Schneider PM (2001) A brief history of the formation of DNA databases in forensic science within Europe. Forensic Sci Int 119(2):225–231
Schneider PM (2009) Expansion of the European standard set of DNA database loci-the current situation. Profile in DNA 2009, 12(1): 6–7. At http://www.promega.com/profiles/1201/1201_06.html
Hares DR (2011) Expanding the CODIS core loci in the United States. Forensic Sci Int Genet 6(1):52–54
Butler JM, Hill CR (2012) Biology and genetics of new autosomal STR loci useful for forensic DNA analysis. Forensic Sci Rev 24:15
Li SL, Liu C, Liu H, Ge JY, Budowle B, Liu CH, Zheng WG, Li FY, Ge BW (2014) Developmental validation of the EX20+4 system. Forensic Sci Int Genet 11:207–213
Shafique M, Shahzad MS, Rahman Z, Shan MA, Perveen R, Shahzad M, Hussain M, Shahid AA, Husnain T (2016) Development of new PCR multiplex system by the simultaneous detection of 10 miniSTRs, SE33, Penta E, Penta D, and four Y-STRs. Int J Legal Med. doi:10.1007/s00414-016-1372-x
Lopes V, Andrade L, Carvalho M, Serra A, Bals F, Bento AM, Batista L, Oliceira C, Corte-Real F, Anjos MJ (2009) Mini-STRs: a powerful tool to identify genetic profiles in samples with small amounts of DNA. Forensic Sci Int Genet Supp S2:121–122
Tenpleton J, Ottens R, Paradiso V, Handt O, Taylor D, Linacre A (2013) Genetic profiling from challenging samples: direct PCR of touch DNA. Forensic Sci Int Genet Supp S4(1):224–225
Zhao G, Zhai H, Yuan Q, Sun S, Liu T, Xie L (2014) Rapid and sensitive diagnosis of fungal keratitis with direct PCR without template DNA extraction. Clin Microbiol Infect 20(10):776–782
Quality Assurance Standards (QAS) for DNA Databasing Laboratories. (2009). Available at: http://www.cstl.nist.gov/strbase/QAS/Final-FBI-Director-Databasing-Standards.pdf
Revised validation guidelines-scientific working group on DNA analysis methods (SWGDAM), Forensic Sci. Commun. 6 (3) (2004), Available at: http://swgdam.org/SWGDAM_Validation_ Guidelines_ APPROVED_Dec_2012.pdf
The implementation of voluntary certification of forensic science product rules—DNA kits, Available at: http://www.csp.gov.cn/UploadFiles/P-V03-005.pdf
Walsh PS, Fildes NJ, Reynolds R (1996) Sequence analysis and characterization of stutter products at the tetranucleotide repeat locus vWA. Nucleic Acids Res 24:2807–2812
Leclair B, Frégeau CJ, Bowen KL, Fourney RM (2004) Systematic analysis of stutter percentages and allele peak height and peak area ratios at heterozygous STR loci for forensic casework and database samples. J Forensic Sci 49(5):968–980
Tereba A (1999) Tools for analysis of population statistics. Profiles DNA 2(3):14–16
Lui K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics 21(9):2128–2129
Weir BS (2007) Matching and partially-matching DNA profiles. Ann Appl Stal 1(2):358–370
Ge JY, Chakraborly R, Eisenberg AJ, Budowle B (2011) Comparisons of familial DNA database searching strategied. J Forensic Sci 56(6):1448–1456
Ge JY, Budowle B, Chakraborly R (2011) Choosing relatives for DNA identification of missing persons. J Forensic Sci 56(s1):23–28
Schneider PM (2007) Scientific standards for studies in forensic genetics. Forensic Sci Int 165(2–3):238–243
Magnuson VL, Ally DS, Nylund SJ, Karanjawala ZE, Rayman JB, Knapp JI, Lowe AL, Ghosh S, Collins FS (1996) Substrate nucleotide-determined non-templated addition of adenine by Taq DNA polymerase: implications for PCR-based genotyping and cloning. BioTechniques 21:700–709
Liu QL, Chen YF, Huang XL, Liu KY, Zhao H, Lu DJ (2016) Population data and mutation rates of 19 STR loci in seven provinces from China based on Goldeneye™ DNA ID System 20A. Int J Legal Med Available at. doi:10.1007/s00414-016-1441-1
Wang Y, Liu C, Zhang CC, Li R, Liu H, Qu XL, Li HX, Sun HY (2016) Analysis of 24 Y-STR haplotype data in a Chinese Han population from Guangdong Province. Int J Legal Med 130(3):689–691
Tozzo P, Giuliodori A, Corato S, Ponzano E, Rodriguez D, Caenazzo L (2013) Deletion of Amelogenin Y-locus in forensics: literature revision and description of a novel method for sex confirmation. J Forensic Legal Med 20(5):387–391
Acknowledgements
This work was supported by a grant from Science and Technology Planning Project of Guangdong Province, China (Grant No. 2013B021500010), Natural Science Foundation of Guangdong Province (Grant No. 2014A030310025), and Medical Science and Technology Research Foundation of Guangdong Province (A2015043).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Weian Du and Ling Chen contributed equally to this work
Rights and permissions
About this article
Cite this article
Du, W., Chen, L., Liu, H. et al. Developmental validation of the HomyGene19+14Y System. Int J Legal Med 131, 605–620 (2017). https://doi.org/10.1007/s00414-016-1505-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00414-016-1505-2