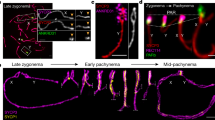
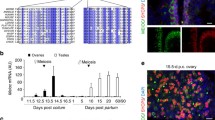
Abstract
Meiosis is a critical phase in the life cycle of sexually reproducing organisms. Chromosome numbers are halved during meiosis, which requires meiosis-specific modification of chromosome behaviour. Furthermore, suppression of transposons is particularly important during meiosis to allow the transmission of undamaged genomic information between generations. Correspondingly, specialized genome defence mechanisms and nuclear structures characterize the germ line during meiosis. Survival of mammalian spermatocytes requires that the sex chromosomes form a distinct silenced chromatin domain, called the sex body. An enigmatic spherical DNA-negative structure, called the meiotic dense body, forms in association with the sex body. The dense body contains small non-coding RNAs including microRNAs and PIWI-associated RNAs. These observations gave rise to speculations that the dense body may be involved in sex body formation and or small non-coding RNA functions, e.g. the silencing of transposons. Nevertheless, the function of the dense body has remained mysterious because no protein essential for dense body formation has been reported yet. We discovered that the polycomb-related sex comb on midleg-like 1 (SCML1) is a meiosis-specific protein and is an essential component of the meiotic dense body. Despite abolished dense body formation, Scml1-deficient mice are fertile and proficient in sex body formation, transposon silencing and in timely progression through meiosis and gametogenesis. Thus, we conclude that dense body formation is not an essential component of the gametogenetic program in the mammalian germ line.








Similar content being viewed by others
References
Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ (2007) Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747
Baarends WM, Wassenaar E, van der Laan R, Hoogerbrugge J, Sleddens-Linkels E, Hoeijmakers JH, de Boer P, Grootegoed JA (2005) Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Mol Cell Biol 25:1041–1053
Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, Burgoyne PS, Jasin M, Keeney S (2005) Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol Cell Biol 25:7203–7215
Baudat F, de Massy B (2007) Regulating double-stranded DNA break repair towards crossover or non-crossover during mammalian meiosis. Chromosom Res 15:565–577
Baudat F, Manova K, Yuen JP, Jasin M, Keeney S (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6:989–998
Beyret E, Lin H (2011) Pinpointing the expression of piRNAs and function of the PIWI protein subfamily during spermatogenesis in the mouse. Dev Biol 355:215–226
Burgoyne PS, Mahadevaiah SK, Turner JM (2009) The consequences of asynapsis for mammalian meiosis. Nat Rev Genet 10:207–216
Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ (2007) MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12:503–514
Cloutier JM, Mahadevaiah SK, ElInati E, Nussenzweig A, Toth A, Turner JM (2015) Histone H2AFX links meiotic chromosome asynapsis to prophase I oocyte loss in mammals. PLoS Genet 11, e1005462
Crackower MA, Kolas NK, Noguchi J, Sarao R, Kikuchi K, Kaneko H, Kobayashi E, Kawai Y, Kozieradzki I, Landers R, Mo R, Hui CC, Nieves E, Cohen PE, Osborne LR, Wada T, Kunieda T, Moens PB, Penninger JM (2003) Essential role of Fkbp6 in male fertility and homologous chromosome pairing in meiosis. Science 300:1291–1295
Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, Roig I, Cooke HJ, Stewart AF, Wassmann K, Jasin M, Keeney S, Toth A (2011) Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol 13:599–610
De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D (2011) The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480:259–263
de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A (2005) Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 19:1376–1389
Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O’Carroll D (2013) Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50:601–608
Dresser ME, Moses MJ (1980) Synaptonemal complex karyotyping in spermatocytes of the Chinese hamster (Cricetulus griseus). IV Light and electron microscopy of synapsis and nucleolar development by silver staining. Chromosoma 76:1–22
Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, Romanienko PJ, Camerini-Otero RD, Bonner WM, Manova K, Burgoyne P, Nussenzweig A (2003) H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev Cell 4:497–508
Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C (2010) A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp Cell Res 316:158–171
Hunter N (2007) Meiotic recombination. In: Molecular genetics of recombination. Springer-Verlag, Berlin Heidelberg
Kargul GJ, Dudekula DB, Qian Y, Lim MK, Jaradat SA, Tanaka TS, Carter MG, Ko MS (2001) Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat Genet 28:17–18
Kim CA, Bowie JU (2003) SAM domains: uniform structure, diversity of function. Trends Biochem Sci 28:625–628
Kogo H, Tsutsumi M, Inagaki H, Ohye T, Kiyonari H, Kurahashi H (2012a) HORMAD2 is essential for synapsis surveillance during meiotic prophase via the recruitment of ATR activity. Genes Cells 17:897–912
Kogo H, Tsutsumi M, Ohye T, Inagaki H, Abe T, Kurahashi H (2012b) HORMAD1-dependent checkpoint/surveillance mechanism eliminates asynaptic oocytes. Genes Cells 17:439–454
Lallemand Y, Luria V, Haffner-Krausz R, Lonai P (1998) Maternally expressed PGK-Cre transgene as a tool for early and uniform activation of the Cre site-specific recombinase. Transgenic Res 7:105–112
Mahadevaiah SK, Bourc’his D, de Rooij DG, Bestor TH, Turner JM, Burgoyne PS (2008) Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J Cell Biol 182:263–276
Marcon E, Babak T, Chua G, Hughes T, Moens PB (2008) miRNA and piRNA localization in the male mammalian meiotic nucleus. Chromosom Res 16:243–260
Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE (2012) AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell 23:251–264
Monesi V (1965) Differential rate of ribonucleic acid synthesis in the autosomes and sex chromosomes during male meiosis in the mouse. Chromosoma 17:11–21
Page SL, Hawley RS (2003) Chromosome choreography: the meiotic ballet. Science 301:785–789
Peters AH, Plug AW, van Vugt MJ, de Boer P (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosom Res 5:66–68
Petronczki M, Siomos MF, Nasmyth K (2003) Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112:423–440
Pillai RS, Chuma S (2012) piRNAs and their involvement in male germline development in mice. Development, growth & differentiation
Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS (2011) Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480:264–267
Romanienko PJ, Camerini-Otero RD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6:975–987
Royo H, Polikiewicz G, Mahadevaiah SK, Prosser H, Mitchell M, Bradley A, de Rooij DG, Burgoyne PS, Turner JM (2010) Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr Biol 20:2117–2123
Royo H, Prosser H, Ruzankina Y, Mahadevaiah SK, Cloutier JM, Baumann M, Fukuda T, Hoog C, Toth A, de Rooij DG, Bradley A, Brown EJ, Turner JM (2013) ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev 27:1484–1494
Royo H, Seitz H, ElInati E, Peters AH, Stadler MB, Turner JM (2015) Silencing of X-linked MicroRNAs by meiotic sex chromosome inactivation. PLoS Genet 11, e1005461
Sabate R, Rousseau F, Schymkowitz J, Ventura S (2015) What makes a protein sequence a prion? PLoS Comput Biol 11, e1004013
Schmid M, Johannisson R, Haaf T, Neitzel H (1987) The chromosomes of Micromys minutus (Rodentia, Murinae). II Pairing pattern of X and Y chromosomes in meiotic prophase. Cytogenet Cell Genet 45:121–131
Shibuya H, Morimoto A, Watanabe Y (2014) The dissection of meiotic chromosome movement in mice using an in vivo electroporation technique. PLoS Genet 10, e1004821
Shoji M, Chuma S, Yoshida K, Morita T, Nakatsuji N (2005) RNA interference during spermatogenesis in mice. Dev Biol 282:524–534
Slot JW, Geuze HJ (2007) Cryosectioning and immunolabeling. Nat Protoc 2:2480–2491
Solari AJ (1974) The behavior of the XY pair in mammals. Int Rev Cytol 38:273–317
Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF (2004) A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis 38:151–158
Tokuyasu KT (1980) Immunochemistry on ultrathin frozen sections. Histochem J 12:381–403
Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37:41–47
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121
Vigodner M, Morris PL (2005) Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev Biol 282:480–492
Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H (2006) Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev 20:1732–1743
Wojtasz L, Daniel K, Toth A (2009a) Fluorescence activated cell sorting of live female germ cells and somatic cells of the mouse fetal gonad based on forward and side scattering. Cytometry A 75A:547–553
Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu HL, Boonsanay V, Eckmann CR, Cooke HJ, Jasin M, Keeney S, McKay MJ, Toth A (2009) Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genet 5
Wojtasz L, Cloutier JM, Baumann M, Daniel K, Varga J, Fu J, Anastassiadis K, Stewart AF, Remenyi A, Turner JMA, Toth A (2012) Meiotic DNA double-strand breaks and chromosome asynapsis in mice are monitored by distinct HORMAD2-independent and -dependent mechanisms. Genes Dev 26:958–973
Wu HH, Su B (2008) Adaptive evolution of SCML1 in primates, a gene involved in male reproduction. BMC Evol Biol 8:192
Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, Benes V, Penninger J, Sachidanandam R, Pillai RS (2012) A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell 47:970–979
Zambrano R, Conchillo-Sole O, Iglesias V, Illa R, Rousseau F, Schymkowitz J, Sabate R, Daura X, Ventura S (2015) PrionW: a server to identify proteins containing glutamine/asparagine rich prion-like domains and their amyloid cores. Nucleic Acids Res 43:W331–W337
Acknowledgments
We thank Rolf Jessberger for sharing ideas, anti-SYCP3 antibody and departmental support, Mary Ann Handel for histone H1T antibody, Nadine Maiwald for the purification of anti-SCML1 antibodies and Daniel Tränkner for lab support and help with Southern blotting. We thank the DIGS-BB program for supporting I.D. CRTD seed grant supported A.G-R. The Deutsche Forschungsgemeinschaft (DFG; grants: SPP1384:TO421/4-1 and 4–2, TO421/3-1, 3–2, 5–1, 6–1, 7–1, 8–1 and 8–2), and MeDDrive 2010 from the TU Dresden Faculty of Medicine supported F.P., K.D., A.G-R., L.W., I.D. and A.T. IMBA, the Austrian Ministry of Sciences, and the Austrian Academy of Sciences supported J.M.P. Wellcome Trust/Cancer Research UK supported A.S. The Max Planck Society supported B.H. An EFRE grant (EuropäischerFondsfürRegionaleEntwicklung) supported J.F. and A.F.S.
Author contributions
Most experiments were carried out by F.P. (Figs. 2, 3b, c, 4, 5, 6, 7, 8, S2 and S4c, e) and K.D. (Figs. 1, 3a, e, S1 and S4d). K.D. identified and cloned the full-length Scml1 open reading frame, generated anti-SCML1 antibodies, devised experiments together with A.T. and contributed to day-to-day supervision of F.P. and A.R. during the initial stages of characterization of Scml1-deficient mice. The Scml1-deficient ES cells and mice were generated by A.G-R. (Fig. S4a, b) with the assistance and supervision of J-F.F. Further experiments were performed by T.K. (Fig. 3d), L.W. (Supplementary Fig. S3b) and I.D. (Supplementary Fig. S3a). Knockout construct was generated by J.F. and A.F.S.; J.P. supplied FKBP6 antibodies and experimental material; A.S. and B.H. provided support for the discovery of Scml1; A.T. wrote the manuscript. All authors were involved in discussions and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All experiments performed in this study involving animals were in accordance with the ethical standards of the Technische Universität Dresden. This article does not contain any studies with human participants performed by any of the authors.
Funding
This work was supported by DFG grants: SPP1384:TO421/4-1 and 4-2, TO421/3-1, 3-2, 5-1, 6-1, 7-1, 8–1 and 8-2, MeDDrive 2010 from the TU Dresden Faculty of Medicine, DIGS-BB program, IMBA, the Austrian Ministry of Sciences, and the Austrian Academy of Sciences, Wellcome Trust/Cancer Research UK, The Max Planck Society and an EFRE grant (EuropäischerFondsfürRegionaleEntwicklung).
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Frantzeskos Papanikos, Katrin Daniel and Angelique Goercharn-Ramlal contributed equally to this work.
Rights and permissions
About this article
Cite this article
Papanikos, F., Daniel, K., Goercharn-Ramlal, A. et al. The enigmatic meiotic dense body and its newly discovered component, SCML1, are dispensable for fertility and gametogenesis in mice. Chromosoma 126, 399–415 (2017). https://doi.org/10.1007/s00412-016-0598-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-016-0598-1