Abstract
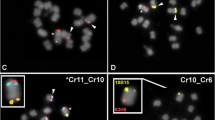
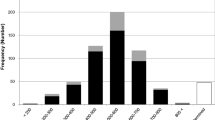
The Indian muntjac (Muntiacus muntjak vaginalis) has a karyotype of 2n=6 in the female and 7 in the male, the karyotypic evolution of which through extensive tandem fusions and several centric fusions has been well-documented by recent molecular cytogenetic studies. In an attempt to define the fusion orientations of conserved chromosomal segments and the molecular mechanisms underlying the tandem fusions, we have constructed a highly redundant (more than six times of whole genome coverage) bacterial artificial chromosome (BAC) library of Indian muntjac. The BAC library contains 124,800 clones with no chromosome bias and has an average insert DNA size of 120 kb. A total of 223 clones have been mapped by fluorescent in situ hybridization onto the chromosomes of both Indian muntjac and Chinese muntjac and a high-resolution comparative map has been established. Our mapping results demonstrate that all tandem fusions that occurred during the evolution of Indian muntjac karyotype from the acrocentric 2n=70 hypothetical ancestral karyotype are centromere–telomere (head–tail) fusions.




Similar content being viewed by others
References
Chang SD, Chao AS, Lai YM, Liu HY, Soong YK (2001) Interphase FISH-assisted second-trimester termination of a trisomy 21 fetus in an IVF-ET twin pregnancy: a case report. J Reprod Med 46:1063–1066
Chi J, Fu B, Nie W, Wang J, Graphodatsky AS, Yang F (2005) New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis). Cytogenet Genome Res 108:310–316
Corbet GB, Hill JE (1992) The mammals of the Indomalayan region: a systematic review. Oxford University Press, New York
Elder FFB, Hsu TC (1988) Tandem fusions in the evolution of mammalian chromosomes. In: Sandberg AA (ed) The cytogenetics of mammalian autosomal rearrangements. Alan R. Liss, New York, pp 481–506
Ferguson-Smith MA (1973) Human autosomal polymorphism and the non-random involvement of chromosomes in translocation. Chromosomes Today 4:235–246
Frönicke L, Scherthan H (1997) Zoo-fluorescence in situ hybridization analysis of human and Indian muntjac karyotypes (Muntiacus muntjak vaginalis) reveals satellite DNA clusters at the margins of conserved syntenic segments. Chromosome Res 5:254–261
Frönicke L, Chowdhary BP, Scherthan H (1997) Segmental homology among cattle (Bos taurus), Indian muntjac (Muntiacus muntjak vaginalis), and Chinese muntjac (M. reevesi) karyotypes. Cytogenet Cell Genet 77:223–227
Hartmann N, Scherthan H (2004) Characterization of ancestral chromosome fusion points in the Indian muntjac deer. Chromosoma 112:213–220
Hsu TC, Pathak S, Chen TR (1975) The possibility of latent centromeres and a proposed nomenclature system for total chromosome and whole arm translocations. Cytogenet Cell Genet 15:41–49
Johnston FP, Church RB, Lin CC (1982) Chromosome rearrangement between the Indian muntjac and Chinese muntjac is accompanied by a deletion of middle repetitive DNA. Can J Biochem 60:497–506
Kehrer-Sawatzki H, Sandig CA, Goidts V, Hameister H (2005) Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet Genome Res 108:91–97
Lee C, Sasi R, Lin CC (1993) Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusion in the karyotypic evolution of the Asian muntjacs. Cytogenet Cell Genet 63:156–159
Li YC, Lee C, Sanoudou D, Hseu TH, Li SY, Lin CC (2000) Interstitial colocalization of two cervid satellite DNAs involved in the genesis of the Indian muntjac karyotype. Chromosome Res 8:363–373
Li YC, Lee C, Chang WS, Li SY, Lin CC (2002) Isolation and identification of a novel satellite DNA family highly conserved in several Cervidae species. Chromosoma 111:176–183
Lin CC, Sasi R, Fan YS, Chen ZQ (1991) New evidence for tandem chromosome fusions in the karyotypic evolution of Asian muntjacs. Chromosoma 101:19–24
Ohtaishi N, Gao Y (1990) A review of the distribution of all species of deer (Tragulidae, Moschidae and Cervidae) in China. Mamm Rev 20:125–144
Qian YP, Jin L, Su B (2004) Construction and characterization of bacterial artificial chromosome library of black-handed spider monkey (Ateles geoffroyi). Genome 47:239–245
Scherthan H (1990) Localization of the repetitive telomeric sequence (TTAGGG)n in two muntjac species and implications for their karyotypic evolution. Cytogenet Cell Genet 53:115–117
Scherthan H (1995) Chromosome evolution in muntjac revealed by centromere, telomere and whole chromosome paint probes. Kew Chromosome Conf IV:267–280
Schmidtke J, Brennecke H, Schmid M, Neitzel H, Sperling K (1981) Evolution of muntjac DNA. Chromosoma 84:187–193
Shi L, Ye YY, Duan XS (1980) Comparative cytogenetic studies on the red muntjac, Chinese muntjac and their F1 hybrids. Cytogenet Cell Genet 26:22–27
Wurster DH, Benirschke K (1967) Chromosome studies in some deer, the springbok and the pronghorn, with notes on placentation in deer. Cytologia 32:273–285
Wurster DH, Benirschke K (1970) Indian muntjac, Muntiacus muntjack: a deer with a low diploid chromosome number. Science 168:1364–1366
Xu HL, Qian YP, Nie WH, Chi JX, Yang FT, Su B (2004) Construction, characterization and chromosomal mapping of bacterial artificial chromosome (BAC) library of Yunnan snub-nosed monkey (Rhinopithecus bieti). Chromosome Res 12:251–262
Yang F (1998) Chromosome evolution of the muntjacs: inferences from molecular cytogenetics. Ph.D. dissertation, University of Cambridge
Yang F, Carter NP, Shi L, Ferguson-Smith MA (1995) A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103:642–652
Yang F, Muller S, Just R, Ferguson-Smith MA, Wienberg J (1997a) Chromosomal evolution of the Chinese muntjac (Muntiacus reevesi). Chromosoma 106:37–43
Yang F, O'Brien PCM, Wienberg J, Ferguson-Smith MA (1997b) A reappraisal of the tandem fusion theory of karyotype evolution in the Indian muntjac using chromosome painting. Chromosome Res 5:109–117
Yang F, O'Brien PCM, Wienberg J, Ferguson-Smith MA (1997c) Evolution of the black muntjac (Muntiacus crinifrons) karyotype revealed by comparative chromosome painting. Cytogenet Cell Genet 76:159–163
Yang F, O'Brien PCM, Wienberg J, Neitzel H, Lin CC, Ferguson-Smith MA (1997d) Comparative chromosome painting in mammals: human and the Indian muntjac (Muntiacus muntjak vaginalis). Genomics 39:396–401
Yang F, Graphodatsky AS, O'Brien PC, Colabella A, Solanky N, Squire M, Sargan DR, Ferguson-Smith MA (2000) Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res 8:393–404
Yue Y, Grossmann B, Ferguson-Smith M, Yang F, Haaf T (2005) Comparative cytogenetics of human chromosome 3q21.3 reveals a hot spot for ectopic recombination in hominoid evolution. Genomics 85:36–47
Acknowledgements
We thank Dr. Charles Lee and two anonymous referees for their critical and constructive review of the manuscript. This research is supported by the National Natural Science Foundation of China (No. 30170506).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E. Nigg
Rights and permissions
About this article
Cite this article
Chi, J.X., Huang, L., Nie, W. et al. Defining the orientation of the tandem fusions that occurred during the evolution of Indian muntjac chromosomes by BAC mapping. Chromosoma 114, 167–172 (2005). https://doi.org/10.1007/s00412-005-0004-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-005-0004-x