Abstract
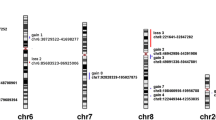
Genomic alterations in lung cancer tissues have been observed in various studies. To analyze the aberrations in the genome of lung cancer patients, we used array comparative genomic hybridization (array CGH) in 15 lung adenocarcinoma (AdC) tissues. Copy number gains and losses in chromosomal regions were detected and corresponding genes were confirmed by real-time polymerase chain reaction (PCR). As for the results, several frequently altered loci, including gain of 16p (46% of samples), were found, and the most common losses were found in 14q32.33 (26% of samples). High-level DNA amplifications (> 0.8 log2 ratio) were detected at 1p, 5p, 7p, 9p, 11p, 11q, 12q, 14q, 16p, 17q, 19q, 20p, 21q, and 22q. A subset of genes, gained or lost, was checked for over- or underrepresentation by means of real-time PCR. The degree of fold change was highest in ECGF1 (22q13.33), HOXA9 (7p15.2), MAFG (17q25.3), TSC2 (16p13.3), and ICAM1 (19p13.2) genes and the 16p chromosome terminal region (16p13.3pter). Taken together, these results show that array CGH could be used as a powerful tool for identification of genomic alteration for lung cancer, and the above-mentioned genes may represent potential candidate genes in the study of lung cancer pathogenesis and diagnosis.


Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Astrinidis A, Khare L, Carsillo T, et al. (2000) Mutational analysis of the tuberous sclerosis gene TSC2 in patients with pulmonary lymphangioleiomyomatosis. J Med Genet 37:55–57
Calvo R, West J, Franklin W, et al. (2000) Altered HOX and WNT7A expression in human lung cancer. Proc Natl Acad Sci USA 97:12776–12781
Chenais B, Derjuga A, Massrieh W, et al. (2005) Functional and placental expression analysis of the human NRF3 transcription factor. Mol Endocrinol 19:125–137
Chujo M, Noguchi T, Miura T, et al. (2002) Comparative genomic hybridization analysis detected frequent overrepresentation of chromosome 3q in squamous cell carcinoma of the lung. Lung Cancer 38:23–29
Garnis C, Lockwood WW, Vucic E, et al. (2005) High-resolution analysis of non-small cell lung cancer cell lines by whole genome tiling path array CGH. Int J Cancer 118:1556–1564
Guillaud-Bataille M, Valent A, Soularue P, et al. (2004) Detecting single DNA copy number variations in complex genomes using one nanogram of starting DNA and BAC-array CGH. Nucleic Acids Res 32:112–120
Hurst CD, Fiegler H, Carr P, et al. (2004) High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene 23:2250–2263
Jiang F, Yin Z, Caraway NP, Li R, Katz RL (2004) Genomic profiles in stage I primary non small cell lung cancer using comparative genomic hybridization analysis of cDNA microarrays. Neoplasia 6:623–635
Mantripragada KK, Buckley PG, de Stahl TD, Dumanski JP (2004) Genomic microarrays in the spotlight. Trends Genet 20:87–94
Mao X, Orchard G, Lillington DM, et al. (2003) Genetic alterations in primary cutaneous CD30+ anaplastic large cell lymphoma. Genes Chromosomes Cancer 37:176–185
Sato J, Sata M, Nakamura H, et al. (2003) Role of thymidine phosphorylase on invasiveness and metastasis in lung adenocarcinoma. Int J Cancer 106: 863–870
Tan D, Deeb G, Wang J, et al. (2003) HER-2/neu protein expression and gene alteration in stage I-IIIA non-small-cell lung cancer: a study of 140 cases using a combination of high throughput tissue microarray, immunohistochemistry, and fluorescent in situ hybridization. Diagn Mol Pathol 12:201–211
Tonon G, Wong KK, Maulik G, et al. (2005) High-resolution genomic profiles of human lung cancer. Proc Natl Acad Sci USA 102:9625–9630
Wikman H, Nymark P, Väyrynen A, et al. (2005) CDK4 is a probable target gene in a novel amplicon at 12q13.3-q14.1 in lung cancer. Genes Chromosomes Cancer 42:193–199
Williams TD, Gensberg K, Minchin SD, Chipman JK (2003) A DNA expression array to detect toxic stress response in European flounder (Platichthys flesus). Aquat Toxicol 65:141–157
Wong MP, Fung LF, Wang E, et al. (2003) Chromosomal aberrations of primary lung adenocarcinomas in nonsmokers. Cancer 97:1263–1270
Yakut T, Egeli U, Gebitekin C (2003) Investigation of c-myc and p53 gene alterations in the tumor and surgical borderline tissues of NSCLC and effects on clinicopathologic behavior: by the FISH technique. Lung 181:245–258
Zhang HW, Zhang L, Chen JH, Du JJ (2005) The clinical significance of vascular endothelial growth factor and intercellular adhesion molecule-1 expression in non-small cell lung cancer. Zhonghua Wai Ke Za Zhi 43:354–357
Zhao X, Weir BA, LaFramboise T, et al. (2005) Homozygous deletions and chromosome amplifications in human lung carcinomas revealed by single nucleotide polymorphism array analysis. Cancer Res 65:5561–5570
Acknowledgments
The authors thank Drs. Wang and Drs. Yun (Department of Thoracic and Cardiovascular Surgery and Department of Internal Medicine, St. Mary’s Hospital, The Catholic University, Korea) for providing the lung tumors from patients. This work was supported by a grant (01-PJ3-PG6-01GN07-0004) from the Korean Health 21 R&D Project, the Ministry of Health and Welfare, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Choi, J.S., Zheng, L.T., Ha, E. et al. Comparative Genomic Hybridization Array Analysis and Real-Time PCR Reveals Genomic Copy Number Alteration for Lung Adenocarcinomas. Lung 184, 355–362 (2006). https://doi.org/10.1007/s00408-006-0009-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00408-006-0009-0