Abstract
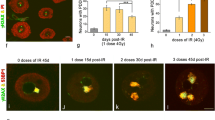
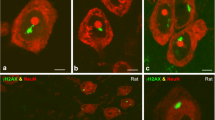
Neurons are very sensitive to DNA damage induced by endogenous and exogenous genotoxic agents, as defective DNA repair can lead to neurodevelopmental disorders, brain tumors and neurodegenerative diseases with severe clinical manifestations. Understanding the impact of DNA damage/repair mechanisms on the nuclear organization, particularly on the regulation of transcription and cell cycle, is essential to know the pathophysiology of defective DNA repair syndromes. In this work, we study the nuclear architecture and spatiotemporal organization of chromatin compartments involved in the DNA damage response (DDR) in rat sensory ganglion neurons exposed to X-ray irradiation (IR). We demonstrate that the neuronal DDR involves the formation of two categories of DNA-damage processing chromatin compartments: transient, disappearing within the 1 day post-IR, and persistent, where unrepaired DNA is accumulated. Both compartments concentrate components of the DDR pathway, including γH2AX, pATM and 53BP1. Furthermore, DNA damage does not induce neuronal apoptosis but triggers the G0–G1 cell cycle phase transition, which is mediated by the activation of the ATM-p53 pathway and increased protein levels of p21 and cyclin D1. Moreover, the run on transcription assay reveals a severe inhibition of transcription at 0.5 h post-IR, followed by its rapid recovery over the 1 day post-IR in parallel with the progression of DNA repair. Therefore, the response of healthy neurons to DNA damage involves a transcription- and cell cycle-dependent but apoptosis-independent process. Furthermore, we propose that the segregation of unrepaired DNA in a few persistent chromatin compartments preserves genomic stability of undamaged DNA and the global transcription rate in neurons.






Similar content being viewed by others
References
Abbas T, Dutta A (2009) p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 9:400–414
Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R (2004) Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science 303:92–95
Baltanas F, Casafont I, Weruaga E, Alonso JR, Berciano MT, Lafarga M (2011) Nucleolar disruption and Cajal body disassembly are nuclear hallmarks of DNA damage-induced neurodegeneration in Purkinje cells. Brain Pathol 21:374–388
Baltanas F, Casafont I, Lafarga V, Weruaga E, Alonso JR, Berciano MT, Lafarga M (2011) Purkinje cell degeneration in pcd mice reveals large scale chromatin reorganization and gene silencing linked to defective DNA repair. J Biol Chem 286:28287–28302
Bauer S, Patterson PH (2005) The cell cycle-apoptosis connection revisited in the adult brain. J Cell Biol 171:641–650
Bekker-Jensen S, Mailand N (2010) Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair 9:1219–1228
Berger SL (2007) The complex language of chromatin regulation during transcription. Nature 447:407–412
Boisvert FM, Lamond AI (2010) p53-dependent subcellular proteome localization following DNA damage. Proteomics 10:4087–4097
Casafont I, Navascués J, Pena E, Lafarga M, Berciano MT (2006) Nuclear organization and dynamics of transcription sites in rat sensory ganglia neurons detected by incorporation of 5′-fluororidine into nascent RNA. Neuroscience 140:453–462
Cavaletti G, Marmiroli P (2010) Chemotherapy-induced peripheral neurotoxicity. Nat Rev Neurol 6:657–666
Chakalova L, Fraser P (2010) Organization of transcription. Cold Spring Harb Perspect Biol 2:a000729
Dellaire G, Kepkay R, Bazett-Jones DP (2009) High resolution imaging of changes in the structure and spatial organization of chromatin, γ-H2AX and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle 8:1–20
Dimitrova N, Chen YC, Spector DL, de Lange T (2008) 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456:524–528
Essers J, Vermeulen W, Houtsmuller AB (2006) DNA damage repair: anytime, anywhere?. Curr Opin Cell Biol 18:240–246
Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A (2004) H2AX: the histone guardian of genome. DNA Repair 3:959–967
Ferrer I, Pozas E, Planas AM (1997) Ubiquitination of apoptotic cells in the developing cerebellum of the rat following ionizing radiation or methylazoxymethanol injection. Acta Neuropathol 93:402–407
Helton ES, Chen X (2007) p53-modulation of the DNA damage response. J Cell Biochem 100:883–896
Hetman M, Vashishta A, Rempala G (2010) Neurotoxic mechanisms of DNA damage: focus on transcriptional inhibition. J Neurochem 114:1537–1549
Iborra FJ, Pombo A, Jackson DA, Cook PR (1996) Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci 109:1427–1436
Khanna KK, Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27:247–254
Klein EA, Assoian RK (2008) Transcriptional regulation of the cyclin D1 gene at a glance. J Cell Sci 121:3853–3857
Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazzett-Jones DP, Nussenzweig A (2006) Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 172:823–834
Kruman II, Wersto RP, Cardozo-Pelaez F, Smilenov L, Chan SL, Chrest FJ, Emokpae R, Gorospe M, Mattson MP (2004) Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron 41:549–561
Kruman II, Schwartz EI (2008) DNA damage response and neuroprotection. Frontiers Biosci 13:2504–2515
Lafarga M, Casafont I, Bengoechea R, Tapia O, Berciano MT (2009) Cajal’s contribution to the knowledge of the neuronal cell nucleus. Chromosoma 118:437–443
Lavin MF (2008) Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signaling and cancer. Nat Rev Mol Cell Biol 9:759–769
Lieber MR (2010) The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Ann Rev Biochem 79:181–211
Ljungman M, Lane DP (2004) Transcription-guarding the genome by sensing DNA damage. Nat Rev Cancer 4:727–737
Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5:255–260
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007) RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900
McKinnon PJ (2009) DNA repair deficiency and neurological disease. Nat Rev Neurosci 10:100–112
Misteli T, Spector DL (2011) The Nucleus. Cold Spring Harbor Lab Press, Cold Spring Harbor
Misteli T, Soutoglou E (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10:243–254
Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O (2007) Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol 178:1101–1108
Noon AT, Shibata A, Rief N, Löbrich M, Stewart GS, Jeggo PA, Goodarzi AA (2010) 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strands break repair. Nature Cell Biol 12:177–184
Nouspikel T (2007) DNA repair in differentiated cells: some new answers to old questions. Neuroscience 145:1213–1221
Olive PL, Banáth JP (2006) The comet assay: a method to measure DNA damage in individual cells. Nat Protocol 1:23–29
Pena E, Berciano MT, Fernandez R, Crespo P, Lafarga M (2000) Stress-induced activation of c-Jun N-terminal kinase in sensory ganglia neurons. Accumulation in nuclear domains enriched in splicing factors and distribution in perichromatin fibrils. Exp Cell Res 256:179–191
Pena E, Berciano MT, Fernandez R, Ojeda JL, Lafarga M (2001) Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comp Neurol 430:250–263
Pryde F, Khalili S, Robertson K, Selfridge J, Ritchie AM, Melton DW, Jullien D, Adachi Y (2005) 53BP1 exchanges slowly at the sites of DNA damage and appears to require RNA for its association with chromatin. J Cell Sci 118:2043–2055
Rass U, Ahel I, West SC (2007) Defective DNA repair and neurodegeneration disease. Cell 130:991–1004
Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3–K9 and H4–K20 trimethylation at constitutive heterochromatin. Genes Dev 18:1251–1262
Shackelford DA (2006) DNA end joining activity is reduced in Alzheimer’s disease. Neurobiol Aging 27:596–605
Shiloh Y (2006) The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci 31:402–441
Shull ERP, Lee Y, Nakane H, Stracker TH, Zhao J, Russell HR, Petrini JHJ, McKinnon PJ (2007) Differential DNA damage signaling accounts for distinct neuronal apoptosis responses in ATLD and NBS. Genes Dev 23:171–180
Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T (2007) Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol 9:675–682
Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Löbrich M, Jeggo PA (2004) ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res 64:2390–2396
Takizawa T, Meshorer E (2008) Chromatin and nuclear architecture in the nervous system. Trends Neurosci 31:343–352
Yang ES, Wang H, Jiang G, Nowsheen S, Fu A, Hallahan DE, Xia F (2009) Lithium-mediated protection of hippocampal cells involves enhancement of DNA-PK-dependent repair in mice. J Clin Invest 119:1124–1135
Acknowledgments
The authors wish to thank Saray Pereda for technical assistance. This work was supported by the following grants: “Dirección General de Investigación” (BFU2008-00175), “Centro de Investigación Biomédica en Red sobre Enfermedades Neurodegenerativas” (CIBERNED; CB06/05/0037) Spain, and “Proyecto I+D+I de la Comunidad de Cantabria” (FMV-UC 09/02) Santander, Spain.
Author information
Authors and Affiliations
Corresponding author
Additional information
I. Casafont and A. Palanca contributed equally to the work reported here.
Rights and permissions
About this article
Cite this article
Casafont, I., Palanca, A., Lafarga, V. et al. Effect of ionizing radiation in sensory ganglion neurons: organization and dynamics of nuclear compartments of DNA damage/repair and their relationship with transcription and cell cycle. Acta Neuropathol 122, 481–493 (2011). https://doi.org/10.1007/s00401-011-0869-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00401-011-0869-0