Abstract
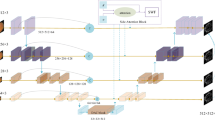
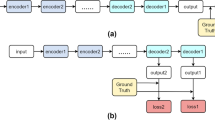
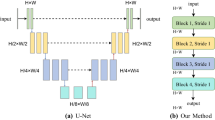
Medical segmentation is a task that pays attention to details. The rapid development of deep learning in image processing technology makes it possible to segment objects accurately on small datasets. In this paper, we propose a hierarchical multi-scale attention network that focuses on the fine-grained parts of the target. Our attention network consists of a hierarchical encoder module with dense connections, a multi-scale module attention to fine-grained parts, and a decoder module. We also combine the weighted cross-entropy loss function based on details and the Dice coefficient loss to increase the sensitivity of fine grains. To verify our module’s performance, we carried out a series of comparative experiments on the multi-scale attention module on the DRIVE dataset. We determine the best structure through experiments and compare it with several classical deep learning models. Our experiments show that extracting semantic information of images at an appropriate resolution can also improve the accuracy of detail segmentation. To show the generalization ability of the work, we conducted experiments on different DRIVE, STARE, and CHASE_DB 1 datasets, and our method achieved 0.8802/0.8464/0.8216 in sensitivity performance metric, 0.9756/0.9869/0.9784 in specificity, and 0.9675/0.9657/0.9637 in accuracy.






Similar content being viewed by others
References
Zhou, S., Nie, D., Adeli, E., Yin, J., Lian, J., Shen, D.: High-resolution encoder–decoder networks for low-contrast medical image segmentation. IEEE Trans. Image Process. 29, 461–475 (2019)
Gibson, E., et al.: Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans. Med. Imaging 37(8), 1822–1834 (2018)
Paulano, F., Jimenez, J.J., Pulido, R.: 3D segmentation and labeling of fractured bone from CT images. Vis. Comput. 30, 939–948 (2014)
Diaz-Pinto, A., Colomer, A., Naranjo, V., Morales, S., Xu, Y., Frangi, A.F.: Retinal image synthesis and semi-supervised learning for glaucoma assessment. IEEE Trans. Med. Imaging 38, 2211–2218 (2019)
Oliveira, A., Pereira, S., Silva, C.A.: Retinal vessel segmentation based on Fully Convolutional Neural Networks. Expert Syst. Appl. 112, 229–242 (2018)
Zhang, J., Chen, Y., Bekkers, E.: Retinal vessel delineation using a brain-inspired wavelet transform and random forest. Pattern Recogn. 69, 107–123 (2017)
Staal, J., Abramoff, M., Niemeijer, M., Viergever, M., Ginneken, B.: Ridge-based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 23(4), 501–509 (2004)
Fu, H., Xu, Y., Lin, S., Kee-Wong, D.W., Liu, J.: DeepVessel: Retinal Vessel Segmentation via Deep Learning and Conditional Random Field, pp. 132–139. Springer, Berlin (2016)
Hafiane, A., Bunyak, F., Palaniappan, K.: Clustering initiated multiphase active contours and robust separation of nuclei groups for tissue segmentation. In: 2008 19th International Conference on Pattern Recognition, pp. 1–4 (2008)
Song, T.H., Sanchez, V., EIDaly, H., Rajpoot, N.M.: Dual-channel active contour model for megakaryocytic cell segmentation in bone marrow trephine histology images. IEEE Trans. Biomed. Eng. 64(12), 2913–2923 (2017)
Lei, B., Kim, J., Kumar, A., Fulham, M., Feng, D.: Stacked fully convolutional networks with multi-channel learning: application to medical image segmentation. Vis. Comput. 33, 1061–1071 (2017)
Fraz, M.M., et al.: An ensemble classification-based approach applied to retinal blood vessel segmentation. IEEE Trans. Biomed. Eng. 59(9), 2538–2548 (2012)
Paulano, F., Jimenez, J.J., Pulido, R.: 3D segmentation and labeling of fractured bone from CT images. Vis. Comput. 30(6–8), 939–948 (2014)
Buyssens, P., Elmoataz, A., Lézoray, O.: Multiscale Convolutional Neural Networks for Vision-Based Classification of Cells, pp. 342–352. Springer, Berlin (2013)
Neverova, N., Wolf, C., Taylor, G.W., Nebout, F.: Multi-Scale Deep Learning for Gesture Detection and Localization, pp. 474–490. Springer, Cham (2015)
Zhuang, J.: LadderNet Multi-Path Networks Based on U-Net for Medical Image Segmentation. arXiv:1810.07810 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Cham, pp. 234–241 (2015)
Badrinarayanan, V., Kendall, A., Cipolla, R.: SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39(12), 2481–2495 (2017)
Angelova, A., Zhu, S.: Efficient object detection and segmentation for fine-grained recognition. In: Presented at the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2013)
Shelhamer, E., Long, J., Darrell, T.: Fully convolutional networks for semantic segmentation. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3431–3440 (2015)
Tu, Z.., Xie, S.: Holistically-nested edge detection. In: The IEEE International Conference on Computer Vision (ICCV), pp. 1395–1403 (2015)
Zhao, H., Shi, J., Qi, X., Wang, X., Jia, J.: Pyramid scene parsing network. In: Presented at the The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017)
Zhao, X., et al.: Fine-Grained Lung Nodule Segmentation with Pyramid Deconvolutional Neural Network (SPIE Medical Imaging). SPIE, San Diego (2019)
Jin, Q., Meng, Z., Pham, T.D., Chen, Q., Wei, L., Su, R.: DUNet: a deformable network for retinal vessel segmentation. Knowl. Based Syst. 178, 149–162 (2019)
Gu, Z., et al.: CE-Net: context encoder network for 2D medical image segmentation. IEEE Trans. Med. Imaging 38, 2281–2292 (2019)
Lecun, Y., Bottou, L., Bengio, Y., Haffner, P.: Gradient-based learning applied to document recognition. Proc. IEEE 86(11), 2278–2324 (1998)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: NIPS, pp. 1097–1105 (2012)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. In: ICLR (2015)
Szegedy, W.L.C., Jia, Y., Sermanet, P., Reed, S.E., Anguelov, D.E.D., Vanhoucke, V., Rabinovich, A.: Going deeper with convolutions. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2015)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2818–2826 (2016)
Lin, G., Milan, A., Shen, C., Reid, I.: RefineNet: Multi-path refinement networks for high-resolution semantic segmentation. In: Presented at the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017)
Chen, L.C., Papandreou, G., Kokkinos, I., Murphy, K., Yuille, A.L.: DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 40(4), 834–848 (2018)
Zhang, X., He, K., Ren, S., Sun, J.: Deep residual learning for image recognition. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778 (2016)
Huang, G., Liu, Z., Der Maaten, L.V., Weinberger, K.Q.: Densely connected convolutional networks. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2261–2269 (2017)
Johnson, J., Cholakkal, H., Rajan, D.: Backtracking ScSPM image classifier for weakly supervised top-down saliency. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 5278–5287 (2016)
Li, Q., Feng, B., Xie, L., Liang, P., Zhang, H., Wang, T.: A cross-modality learning approach for vessel segmentation in retinal images. IEEE Trans. Med. Imaging 35(1), 109–118 (2016)
Orlando, J.I., Prokofyeva, E., Blaschko, M.B.: A discriminatively trained fully connected conditional random field model for blood vessel segmentation in fundus images. IEEE Trans. Biomed. Eng. 64(1), 16–27 (2017)
Alom, M.Z., Hasan, M., Yakopcic, C., Taha, T.M., Asari, V.K.: Recurrent Residual Convolutional Neural Network based on U-Net (R2U-Net) for Medical Image Segmentation. arXiv preprint arXiv:1802.06955 (2018)
Lei, B., Kim, J., Kumar, A., Fulham, M., Feng, D.: Stacked fully convolutional networks with multi-channel learning: application to medical image segmentation. Vis. Comput. 33(6), 1061–1071 (2017)
Wang, D., Haytham, A., Pottenburgh, J., Saeedi, O., Tao, Y.: Hard attention net for automatic retinal vessel segmentation. IEEE J. Biomed. Health Inform. 2020, 1 (2020)
Wu, Y., Xia, Y., Song, Y., Zhang, Y., Cai, W.: NFN+: A novel network followed network for retinal vessel segmentation. Neural Netw. 126, 153–162 (2020)
Fang, L., Wang, C., Li, S., Rabbani, H., Chen, X., Liu, Z.: Attention to lesion: lesion-aware convolutional neural network for retinal optical coherence tomography image classification. IEEE Trans. Med. Imaging 38(8), 1959–1970 (2019)
Liu, Z., Li, X., Luo, P., Change Loy, C., Tang, X.: Not all pixels are equal difficulty-aware semantic segmentation via deep layer cascade. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3193–3202 (2017)
Yan, Z., Yang, X., Cheng, K.T.: A three-stage deep learning model for accurate retinal vessel segmentation. IEEE J. Biomed. Health Inform. 23(4), 1427–1436 (2019)
Owen, C.G., Rudnicka, A.R., Mullen, R., Barman, S.A., Monekosso, D., Whincup, P.H., et al.: Measuring retinal vessel tortuosity in 10-year-old children: validation of the computerassisted image analysis of the retina (CAIAR) program. Investig. Ophthalmol. Vis. Sci. 50, 2004–2010 (2009)
Acknowledgements
This work is supported by the Nature Science Foundation of Guangdong province, No. 2016A030313520.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest in the submission.
Ethical standards
We would like to state that this work is original research that has not been published before and is not considered for publication elsewhere.
Humans and animal rights
The paper does not include any studies of humans or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lyu, C., Hu, G. & Wang, D. Attention to fine-grained information: hierarchical multi-scale network for retinal vessel segmentation. Vis Comput 38, 345–355 (2022). https://doi.org/10.1007/s00371-020-02018-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-020-02018-w