Abstract
Purpose
To established an AI system to make the pathological diagnosis of prostate cancer.
Methods
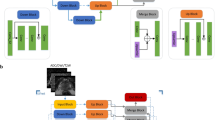
Prostate histopathological whole mount (WM) sections from patients underwent robot-assisted laparoscopic prostatectomy were prepared. All the prostate WM pathological sections were converted to digital image data and marked with different colors on the basis of the ISUP Gleason grade group. The image was then fed into a segmentation algorithm. We chose modified U-Net as our fundamental network architecture.
Results
172 patients were involved in this study. 896 pieces of prostate WM pathological sections from 160 patients, in which 826 pieces of WM sections from 148 patients were assigned to the training set randomly. After image segmentation there were totally 2,138,895 patches, of which 1,646,535 patches were valid for training. The other WM section was arranged for testing. Based on the whole image testing, AI and pathologists presented the same answers among 21 of 22 pieces of sections. To evaluate the diagnostic results at the pixel level, we anticipated correct cancer or non-cancer diagnose from this AI system. The area under the ROC curve as 96.8%. The value of pixel accuracy of three methods (binary analysis, clinically oriented analysis and analysis for different ISUP Gleason grade) were 96.93%, 95.43% and 93.88%, respectively. The value of frequency weighted IoU were 94.32%, 92.13% and 90.21%, respectively.
Conclusions
This AI system is able to assist pathologists to make a final diagnosis, indicating the great potential and a wide-range of applications of AI in the medical field.





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author, HG, upon reasonable request.
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics, 2018. CA Cancer J Clin 68(1):7–30. https://doi.org/10.3322/caac.21442
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 15:10. https://doi.org/10.3322/caac.21492
Rice KR, Furusato B, Chen Y, McLeod DG, Sesterhenn IA, Brassell SA (2009) Clinicopathological behavior of single focus prostate adenocarcinoma. J Urol 182(6):2689–2694. https://doi.org/10.1016/j.juro.2009.08.055
Epstein JI, Zelefsky MJ, Sjoberg DD, Nelson JB, Egevad L, Magi-Galluzzi C et al (2016) A contemporary prostate cancer grading system: a validated alternative to the Gleason score. Eur Urol 69(3):428–435. https://doi.org/10.1016/j.eururo.2015.06.046
Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson AN et al (2015) Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA 313(11):1122–1132. https://doi.org/10.1001/jama.2015.1405
Al-Maghrabi JA, Bakshi NA, Farsi HM (2013) Gleason grading of prostate cancer in needle core biopsies: a comparison of general and urologic pathologists. Ann Saudi Med 33(1):40–44. https://doi.org/10.5144/0256-4947.2013.40
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on neural information processing systems—Volume 1. Lake Tahoe, Nevada: Curran Associates Inc., pp 1097–105
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al (2015) Going deeper with convolutions. In: Cvpr
Liu S, Deng W (2015) Very deep convolutional neural network based image classification using small training sample size. In: 2015 3rd IAPR Asian Conference on pattern recognition (ACPR), pp 730–4.
He K, Zhang X, Ren S, Sun J (2015) Deep residual learning for image recognition. CoRR abs/1512.03385 (2015)
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: 2015 IEEE Conference on computer vision and pattern recognition (CVPR), pp 3431–40
Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL (2016) DeepLab: semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs (2016). arXiv preprint arXiv:160600915
Dai J, Li Y, He K, Sun J (2016) R-fcn: Object detection via region-based fully convolutional networks. In: Advances in neural information processing systems, pp 379–87
Ren S, He K, Girshick R, Sun J (2015) Faster r-cnn: Towards real-time object detection with region proposal networks. In: Advances in neural information processing systems, pp 91–9
Rabilloud N, Allaume P, Acosta O, De Crevoisier R, Bourgade R, Loussouarn D et al (2023) Deep learning methodologies applied to digital pathology in prostate cancer: a systematic review. Diagnostics (Basel). https://doi.org/10.3390/diagnostics13162676
Frewing A, Gibson AB, Robertson R, Urie PM, Corte DD (2023) Don’t fear the artificial intelligence: a systematic review of machine learning for prostate cancer detection in pathology. Arch Pathol Lab Med. https://doi.org/10.5858/arpa.2022-0460-RA
McNeal JE, Haillot O (2001) Patterns of spread of adenocarcinoma in the prostate as related to cancer volume. Prostate 49(1):48–57. https://doi.org/10.1002/pros.1117
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) UNet++: a nested U-NET architecture for medical image segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support 2018(11045):3–11. https://doi.org/10.1007/978-3-030-00889-5_1
Obuchowski NA (1997) Nonparametric analysis of clustered ROC curve data. Biometrics 53(2):567–578
Garcia-Garcia A, Orts-Escolano S, Oprea S, Villena-Martinez V, Garcia-Rodriguez J (2017) A review on deep learning techniques applied to semantic segmentation. arXiv preprint arXiv:170406857
Litjens G, Sanchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I et al (2016) Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep 6:26286. https://doi.org/10.1038/srep26286
Yin Y, Zhang Q, Zhang H, He Y, Huang J (2017) Molecular signature to risk-stratify prostate cancer of intermediate risk. Clin Cancer Res 23(1):6–8. https://doi.org/10.1158/1078-0432.CCR-16-2400
Kayalibay B, Jensen G, van der Smagt P (2017) CNN-based segmentation of medical imaging data. arXiv preprint arXiv:170103056
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234–41
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284(2):574–582. https://doi.org/10.1148/radiol.2017162326
Norman B, Pedoia V, Majumdar S (2018) Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR IMAGING DATA TO DETERMINE RELAXOMETRY AND MORPHOMEtry. Radiology 288(1):177–185. https://doi.org/10.1148/radiol.2018172322
Acknowledgements
This study was funded by the National Natural Science Foundation of China (81902581, 81572519, 81772710), Basic Research Program (Natural Science Foundation) of Jiangsu Province (BK20190117), China Postdoctoral Foundation (2019M660111), Mobility Programme of Sino-German Center (M-0670)
Author information
Authors and Affiliations
Contributions
CZ: Project development, Data collection, Data analysis, Manuscript writing. XG: Data analysis, Manuscript writing. BF: Data collection, Manuscript writing. SG: Data analysis, Manuscript writing. XL: Data collection. JS: Data analysis. YF: Data analysis. QZ: Project development, Manuscript editing. PL: Project development. HG: Project development.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Anonymous data of human participants were involved in this study. The study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki. Animals were not included in this study.
Informed consent
All the patients involved in the study were provided signed informed consent, which was approved by the ethics committee of Nanjing Drum Tower Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Gao, X., Fan, B. et al. Highly accurate and effective deep neural networks in pathological diagnosis of prostate cancer. World J Urol 42, 93 (2024). https://doi.org/10.1007/s00345-024-04775-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00345-024-04775-y