Abstract
Despite the marine environment being typified by a lack of obvious barriers to dispersal, levels of genetic divergence can arise in marine organisms from historical changes in habitat availability, current oceanographic regimes and anthropogenic factors. Here we describe the genetic structure of the Gray Parrotfish, Sparisoma axillare, and identify environmental variables associated with patterns of genetic divergence throughout most of its distribution in Brazil. The heavily exploited Gray Parrotfish is endemic to Brazil, and there is lack of data on population structure that is needed to support sustainable management. To address this shortfall we analyzed 5429 SNPs from individuals sampled in nine locations, ranging from tropical to subtropical reef systems and costal to oceanic environments with varying levels of protection. We found low levels of genetic structure along the coast, including the oceanic island of Fernando de Noronha, and that a combination of water depth, ocean currents and geographic distance were the major drivers explaining genetic divergence. We identified a distinct genetic population around Trindade Island, 1000 km from the coast, highlighting the conservation significance of this population. Colonization of this oceanic site probably occurred during the Pleistocene periods of lower sea levels, allowing this shallow water-dependent species to use the seamount chain as stepping stones to Trindade. Our data further suggest that two protected areas, Costa dos Corais and Fernando de Noronha, likely play an important role as larval sources for much of the species distribution.
Similar content being viewed by others
Introduction
Barriers to the dispersal and gene flow of organisms are usually less obvious in marine than in terrestrial environments, and there are inherent challenges in measuring dispersal for many marine organisms. This had led to relatively less information on population structuring in the marine environment, even for commercially exploited species. Although it may be assumed that widespread gene flow is more common place for marine organisms, genetic discontinuities and speciation are more common than might be expected (Rocha et al. 2007), thereby implicating physical and ecological influences (Lundy et al. 1999; Baker et al. 2008; Cunningham et al. 2009; Roy et al. 2012; Bradbury et al. 2013; Albaina et al. 2013; Truelove et al. 2017; Córdova-Alarcón et al. 2019). For example, even though the pelagic larval phase present in many marine organisms is largely dispersed by currents, how this shapes genetic structure is variable (Gilg and Hilbish 2003; Johnson and Black 2006; Weersing and Toonen 2009; Luiz et al. 2013). Currents can either augment self-recruitment patterns or decrease the local dispersal of a species by carrying larvae away from natal reefs (Treml et al. 2008). Species with a pelagic larval phase may be sedentary in later life stages; therefore, dispersal is mostly dictated by passive movement during pelagic larval stages (Victor and Wellington 2000; Mora and Sale 2002). The duration of the pelagic larval phase along with the geographic distribution of suitable habitat further influences the scale at which dispersal and gene flow occur (now collectively referred to as ‘connectivity’), with shorter larval phases generally associated with geographic isolation seen at finer spatial scales (Saenz-Agudelo et al. 2015; Pinheiro et al. 2017; Riginos and Nachman 2001; Dixo et al. 2009; Luiz et al. 2012).
The Brazilian Province is emerging as a region of intense study of biogeography of marine organisms (e.g., Floeter et al. 2008; Liedke et al. 2020; Simon et al. 2021). A major feature shaping the distribution of marine organisms and endemism in the Province is ocean currents and ecological barriers. These include the Amazon-Orinoco Plume that affects the degree of connectivity with the Caribbean biodiversity and the Mid Atlantic Barrier that filters migration with the Eastern Atlantic (Floeter et al. 2008; Luiz et al. 2012). The reef fish fauna within the Brazilian Province was also shaped by periodic changes in connectivity, with a combination of the Amazon plume effect and historical variation in sea level influencing the degree of isolation from the Caribbean (Rocha 2003; Volk et al. 2021). Levels of connectivity were also intermittent with the East Atlantic (Luiz et al. 2012), allowing occasional biodiversity exchange and gene flow from Africa and the Indo-Pacific (Sánchez and Alvarez 1988; Rocha et al. 2005; Bowen et al. 2006).
In addition to past changes in sea level, the continental shelf width also influences levels of connectivity in estuarine species, with a broader continental shelf in the south of the Brazilian Province sustaining higher levels of gene flow among populations (e.g., Atherinella brasiliensis-Baggio et al. 2017). This is because a wider continental shelf provides more extensive areas that allow for the expansion of coastal habitats used by estuarine species during sea level change (Baggio et al. 2017). Additionally, riverine discharge and sharp changes in temperature due to regional upwelling events also play a role in generating genetic divergence of fish (Santos et al. 2006; Machado et al. 2017; Dias et al. 2019; Volk et al. 2021) and invertebrates (Guo et al. 2015; Strugnell et al. 2017; Takeuchi et al. 2020). The biogeographic scenario established by history, the ecological gradients and different oceanographic characteristics within this Province provides a complex system to explore the processes associated with the gene flow and diversification for many taxa (Beheregaray et al. 2002; Santos et al. 2006; Nunes et al. 2011; Silva et al. 2016; Pinheiro et al. 2017; Marceniuk et al. 2019).
To explore how ecological features affect gene flow and genetic divergence, the taxa researched need to be distributed across heterogeneous habitats (Rocha et al. 2007). Here we focus on a species belonging to the tribe of parrotfishes Sparisomatini, a group that is endemic to the Atlantic and have an origin in the Miocene. This group includes browser and herbivorous-microphages (scraper and excavator) species, radiating to inhabit a range of reef habitats (Ferreira and Gonçalves 2006; Clements et al. 2017; Siqueira et al. 2019). Seven Sparisoma species are distributed in rocky and coral reefs along the Brazilian Province, with four endemic species (Pinheiro et al. 2018). Sparisoma parrotfishes are an excellent model to assess genetic connectivity, given that they are widespread and mostly common in both coastal regions and oceanic islands (Mazzei et al. 2019, 2021). The Amazonas outflow appears to be a major ecological barrier for this group, splitting lineages between Caribbean and Brazilian parrotfishes as demonstrated by allopatric speciation of the Redfin Parrotfish (Sparisoma rubripinne) from the Caribbean and the Gray Parrotfish (Sparisoma axillare) in the Brazilian Province (Robertson et al. 2006). Sparisoma axillare is endemic to Brazil and is a relatively abundant species in shallow coastal and oceanic reef habitats (Ferreira et al. 2004; Roos et al. 2015; Longo et al. 2019). Its pelagic larval phase is unknown, but it is assumed to be similar to other Sparisoma species (average between 57 and 60 d; Robertson et al. 2006). Moreover, it can occupy different habitats throughout its ontogenetic development (Feitosa and Ferreira 2015; Eggertsen et al. 2017; Roos et al. 2019). Sparisoma axillare has a relatively large body size (40 cm standard length) and is a target of fishing from the north to southeastern Brazilian coast (Bender et al. 2014; Roos et al. 2015; Barbosa et al. 2021). Consequently, populations have declined and S. axillare has been classified as ‘vulnerable’ in the Brazilian List of Endangered Aquatic Animals (MMA 2022), requiring management and conservation measures, such as catch limits and the assignment of Marine Protected Areas (MPAs). Biophysical models of Sparisoma larvae along the Brazilian Province have predicted low larval connectivity between the existing MPAs, and few recruitment spots are protected (Endo et al. 2019).
Given the wide distribution of the endemic S. axillare across Brazilian reefs, the level of exploitation and general threats to reef habitats, it is valuable to characterize the genetic structure of this species in order to guide management and conservation actions. A recently published recovery plan that focused on parrotfishes in Brazil, including S. axillare, suggested that exploitation of these species should only occur inside IUCN category IV protected areas (a category that allows a sustainable level of exploitation) under specific rules, and be banned elsewhere (Pinheiro et al. 2021). In light of this, knowledge of gene flow between protected and unprotected areas can assist managing for population sustainability. Further, the patterns of genetic structure for this species may represent a proxy for co-occurring reef species that share similar life histories. To provide this information we used single nucleotide polymorphisms (SNPs) to characterize the genetic structure and identify the environmental variables that are associated with the genetic divergence of S. axillare populations for most of its distribution along the Brazilian Province.
Materials and methods
Study area
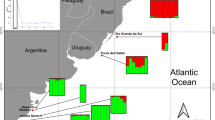
Brazil’s extensive coastline together with its continental shelf and Exclusive Economic Zone (EEZ) encompasses a wide diversity of habitats that include coastal and oceanic islands with different origins, features and distances from the coast, and which are affected differently by ocean currents (Ferreira et al. 2004; Floeter et al. 2008; Pinheiro et al. 2018). Samples were collected along more than 4,000 km of the Brazilian coast, including seven coastal sites, and three off-coast sites (two oceanic islands and one mid-shelf reef site; Fig. 1, Table S1). The northeast coastal sites—Rio Grande do Norte (RN), Pernambuco (PE), Alagoas (AL)—are in regions with tropical waters and narrow continental shelf. The central coastal sites—Bahia (BA) and Espírito Santo (ES)—are in a wider continental shelf. The southernmost coastal site, Rio de Janeiro (RJ), is also in a wider continental shelf and has lower sea temperatures. The three off-coast sites include the volcanic oceanic archipelagos of Fernando de Noronha (FNA) and Trindade (TRI), and the mid-shelf reef of Abrolhos (ABR), the last one located inside the continental shelf. FNA, about 400 km off RN coast, is connected by a chain of seamounts and shares multiple features with the coast, including most of its fish fauna (Floeter et al. 2008; Pinheiro et al. 2018). TRI is located 1100 km off the ES coast and is connected to the coast through a chain of 30 volcanic seamounts, the Vitoria-Trindade Chain (VTC; Fig. 1). The sites that have a low level of environmental protection are PE and AL (coastal MPA of Costa do Corais) and the three off-coast sites.
Sampling sites (red dots), the coastal sites: RN—Rio Grande do Norte, PE—Pernambuco, AL—Alagoas, BA—Bahia, ES—Espírito Santo, RJ—Rio de Janeiro, and islands sites are: FNA—Fernando de Noronha, ABR—Abrolhos Bank, TRI—Trindade Island, and the location of the coastal Marine Protected Area of Costa dos Corais is represented by a triangle
Sampling
Eighty-nine S. axillare tissue samples (fin clips) were collected between 2017 and 2018 at ten sites by SCUBA diving (SISBIO authorization number 48112-7) and from fish landings and fish markets (when the fish origin was known). Tissue samples were stored in 95% ethanol for later analysis. The number of samples collected per site ranged between six and twelve (except for the samples from ES, which consisted of only four, Supplementary Material Table S1).
SNP sequencing and filtering
DNA extraction and SNP discovery were performed by Diversity Arrays Technology Pty. Ltd. (DArT). Initially SNPs were identified and genotyped using the DArTseq method, which is similar to ddRad-seq and based on a combination of complexity reduction and next-generation sequencing methods (described by Jaccoud et al. 2001). Restriction enzymes were selected specifically for parrotfish by testing several enzyme combinations. The resulting fragments were amplified using PCR and sequenced using Illumina Hiseq2500. To exclude data resulting from cross-contamination, the NCBI database was searched for sequence matches with bacterial, fungal and human sequences using BLASTn. All remaining sequences were analyzed using DArTsoft14 software to call SNPs (Jaccoud et al. 2001). Analysis of controlled cross-populations (~ 1000) was performed. To guarantee that true allelic variants from the paralogous sequences were called, the populations were tested for Mendelian distribution of the alleles. The resulting list of SNPs was then provided by DArT, along with information for each marker, such as average reproducibility, i.e., the reproducibility fraction of each of the two alleles and averaged for each marker, which was present in around 30% of technical replication results. This dataset was filtered further using the dartR package (Gruber et al. 2018) in R 3.5 (R Core Team 2019) using the following parameters: minimum call rate of 95% (gl.filter.callrate function) and 100% average reproducibility (gl.filter.reproducibility function). When more than one SNP was found in the same fragment, the SNP with the highest frequency was retained to avoid physically linked loci (gl.filter.secondaries function, applying method = ‘best’). A single individual with more than 20% of missing data was excluded. After following these filtering steps, 5,429 SNPs and 88 individuals were kept for the subsequent analyses.
Genetic structure
To characterize genetic structure for S. axillare pairwise, FST was estimated and three complementary genetic clustering analyses were applied (PCA, DAPC and STRUCTURE) to test whether a consistent pattern emerges despite the varying assumptions with each of these approaches. FST between sampling sites was estimated in R using the function gl.fst.pop from the dartR package. A principal component analysis (PCA) was used to visually inspect population structure according to genetic similarities (Jombart 2008). The first part of this analysis aimed to detect the proportion of variance explained by each principal component (PC). After choosing the number of PCs that explained more than 80% of the variation, a PCA was run using the glPca function of the adegenet package (Jombart 2008) and results plotted using the gl.pcoa.plot function in R. A Discriminant Analysis of Principal Components (DAPC) was carried out with the dapc function of the adegenet package in R to provide an estimate of genetic structure that does not assume that individual genetic clusters approximate Hardy–Weinberg equilibrium (Jombart et al. 2010). DAPC uses ordination to graphically describe the genetic variation. Whereas PCA maximizes the similarity within groups, DAPC maximizes the divergence between groups and reduces genetic divergence within groups. Depending on the migration pattern, DAPC can be more effective at identifying the real number of clusters than other analyses (Jombart et al. 2010). Initially, the function find.clusters was used to detect the optimal number of clusters, by testing from k = 1 to k = 9, with 105 iterations, and the best number was defined based on the lower BIC (Bayesian information criteria) (Jombart et al. 2010). To test for population structure without including a priori information about sampling locations, the STRUCTURE Software v2.3.4 was used (Pritchard et al. 2000). This software uses a Bayesian iterative algorithm and clusters individuals into groups based on allele frequency data using the best fit and the lowest deviation from Hardy–Weinberg and linkage equilibrium. The parameters applied in STRUCTURE analysis were as follows: to test from 1 to 9 possible clusters (10 independent iterations each), to use the Admixture model, to consider the allele frequencies of each population to be dependent and to have a run length of 100,000 (10,000 burn-in). The most probable number of K (number of clusters) was inferred based on ΔK (Evanno et al. 2005) and on the likelihood (Pritchard et al. 2000).
Isolation by distance and resistance
To test for isolation by distance the geographical distances between sampling sites were measured in Google Earth in kilometers. Genetic similarity between sampling sites was calculated using the function dist.genpop from adegenet package in R. To test the significance of any association between the log of geographic distance and genetic similarity, a Mantel test was applied using the function mantel.randtest from ade4 package in R (Dray and Dufour 2007).
To test the influence of environmental factors on genetic divergence the resistance to dispersal was evaluated for four variables: bathymetry (m), mean velocity of currents (m−1), minimum sea surface temperature (°C) and mean terrain curvature of seafloor (°). Bathymetry can affect connectivity given that deep water provides unsuitable habitat for S. axillare (Roos et al. 2019). Minimum sea surface temperature was used given that S. axillare occurs mostly in warm waters (Pinheiro et al. 2018) and, thus, regions with colder waters could act as barriers. Curvature, which is derived from the depth of the seafloor, indicates how bottom currents can interact with the seafloor and, potentially, in turn affect dispersal. Marine currents were tested because currents can contribute toward connectivity by carrying the pelagic larvae of this species.
Variables were extracted from both bio-oracle (Tyberghein et al. 2012; Assis et al. 2018) and MARSPEC (Sbrocco and Barber 2013) and how the resistance values attributed to each variable were categorized are shown in Table S2 (Supplementary Material). To define the environmental distance between sites, the cumulative resistance between sites was calculated using the Circuitscape software (McRae and Beier 2007) and the ResistanceGA package (Peterman 2018) for each variable in R. Circuitscape is based on the circuit theory approach to identify all possible pathways between sites (focal points). Initially, to test for correlations between environmental distances and genetic divergence, Mantel tests were applied using the same approach applied for IBD analysis. To further evaluate significantly associated environmental variables (those with p-values lower than 0.05) while accounting for geographical distance, maximum likelihood population effects (MLPE) models were tested using the ResistanceGA package (Clarke et al. 2002; Peterman 2018). The MLPE models were then ranked by AICc using the function model.sel in R. The best models were those with lower AICc values and higher AIC weights.
Results
Genetic population structure
Pairwise FST between Sparisoma axillare collection sites ranged from 0.01 (FNA and PE) and 0.175 (ABR and TRI) (Supplementary Material Table S3), and global FST was ~ 0.02 and significantly different from zero. The coastal locations with significant pairwise FST, were RN and AL. Divergence between PE and most of the other sites was low, as indicated by the either low or nonsignificant FST values. The off-coast sites FNA and ABR had low pairwise FST with the coastal sites. When comparing these two off-coast sites with AL, the FST was low but significantly different from zero. The FST between the third off-coast site, TRI, and all the other sites, including the other off-coast sites, were always significant and higher than 0.1.
The PCA corroborated the FST results, which showed greater genetic differences between TRI and all the other sites (Fig. 2). PC1 showed a clear divergence between TRI and all the other populations, whereas PC2 indicated some level of divergence between two individuals from ABR and the other sites. PC1 and PC2 explained 6.2% and 2.3% of the total variance, respectively (Fig. 2). The divergence between two other individuals from ABR and all the other sites is shown in PC3 and explains an additional 2.1% of the genetic variance (Supplementary Material Fig. S1).
Principal component analysis (PCA) of genomic similarity based on 5429 Sparisoma axillare SNPs from nine sites located on Brazilian coast and islands: RN—Rio Grande do Norte, PE—Pernambuco, AL—Alagoas, BA—Bahia, ES—Espírito Santo, RJ—Rio de Janeiro, FNA—Fernando de Noronha, ABR—Abrolhos Bank and TRI—Trindade Island
The BIC indicated that the most likely number of clusters is 2, thus suggesting two divergent populations. The DAPC analysis suggested a similar pattern to that found in the first PC of the PCA, thus showing that there is a divergence between the TRI population and all the other sites (Supplementary Material Fig. S2a). Another DAPC analysis excluding TRI estimated that the most probable number of clusters was 1 (Supplementary Material Fig. S2b).
The STRUCTURE analysis shows that K = 2 is the best model (Fig. 3). Again, one cluster comprised all the coastal sites plus ABR and FNA, and the other comprised only TRI. The barplot for K = 3 and K = 4 provides some evidence for the genetic differentiation of ABR, FNA and AL (Fig. 3). We also run STRUCTURE excluding the most divergent site, TRI, to avoid masking smaller divergences. Results also indicated that all the other sampled sites most likely comprise a single cluster (Supplementary Material Fig. S3).
Barplot of the STRUCTURE analysis showing A) the most likely number of clusters of Sparisoma axillare (K = 2), B) K = 3, C) K = 4: the genetic characteristics associated with the Trindade Island (TRI) population are indicated by the color blue, and all the other sites are indicated by the colors pink, yellow and gray depending on the number of populations being tested. Each vertical bar represents one individual, and the color indicates the probability of each individual in each ancestral cluster. The TRI cluster (blue) shows that all the individuals sampled in TRI are 100% confined to TRI at K = 2 (the best supported model). The sampling locations are abbreviated as follows; TRI—Trindade Island, FNA—Fernando de Noronha Archipelago, ABR—Abrolhos Bank, RN—Rio Grande do Norte, PE—Pernambuco, AL—Alagoas, BA—Bahia, ES—Espírito Santo, RJ—Rio de Janeiro
Isolation by distance and resistance
Isolation by geographic distance was not significant for either the complete dataset (p = 0.088) or the dataset excluding TRI (p = 0.196; Fig. 4a). Analyses of environmental distance via Mantel testing resulted in a single significant association between bathymetry with genetic distance when TRI was included (p = 0.025, Fig. 4b). When comparing the models (i) genetic divergence ~ bathymetry, (ii) genetic divergence ~ geographic distance and (iii) genetic divergence ~ bathymetry + geographic distance, the lowest AIC indicates that (i) bathymetric resistance best explained the genetic distance, followed by (iii) the combination of geographic distance and bathymetry (Supplementary Material Table S4).
Isolation by a geographic distance and b environmental resistance (bathymetry) for Sparisoma axillare populations in Brazil. Genetic distance was calculated using genetic dissimilarity (Euclidian). Circles indicate pairwise comparison between coastal sites. Squares indicate pairwise comparison between one off-coast site and one coastal site. Triangles indicate pairwise comparison between off-coast sites. All the points on the top right of both graphs are comparison between Trindade Island (TRI) and all the other sites. Circles in yellow and orange are pairwise comparisons between coastal sites that are located geographically farther away, with the same or higher distances than TRI and the other sites, but with lower genetic divergence. TRI—Trindade Island, FNA—Fernando de Noronha Archipelago, ABR—Abrolhos Bank, RJ—Rio de Janeiro, ES—Espírito Santo, BA—Bahia, PE—Pernambuco, RN—Rio Grande do Norte. “Coast” includes the following sites: RJ, ES, BA, AL (Alagoas), PE and RN
Discussion
We characterized the genetic structure of the endemic and vulnerable Gray Parrotfish Sparisoma axillare across most of its distribution in Brazilian waters. We revealed that populations of S. axillare are connected to varying degrees, and that genetic divergence is largely a consequence of water depth. The off-coast TRI was consistently identified as genetically differentiated and inferred to be the most isolated, most likely due to the geographic extent of deep water separating this population with the rest of the distribution. In contrast, the FNA off-coast site showed a high level of connectivity with most coastal sites, most likely a consequence of oceanographic characteristics. FST analyses and structure analyses also detected significant but low levels of genetic differentiation, even between sites in relatively close geographic proximity. The extent of genetic divergence was unexpected given that S. axillare has a pelagic larval phase, a trait that often confers relatively high rates of dispersal.
The high genetic differentiation between the S. axillare population at TRI and other sites is consistent with studies of corals (Peluso et al. 2018) and other fishes (Simon et al. 2021), although contrasting results have been reported for some other taxa (Teschima et al. 2016; Liedke et al. 2020; Simon et al. 2021). This genetic divergence can be explained by a combination of the large geographic distance and the lack of suitable shallow habitat between TRI and the closest coastal site (~ 1000 km). Although the relevance of bathymetry to genetic divergence could become more pronounced with increasing geographic distance, our data indicate that geographic distance alone does not significantly explain the genetic structure.
Historically, glacial cycles likely affect the levels of dispersal and gene flow, with increased gene flow between the Brazilian Province and Caribbean during sea level rise (Rocha 2003; Ludt and Rocha 2015). In contrast, the connectivity between the coast and TRI is likely to increase as the sea level drops (Pinheiro et al. 2017; Simon et al. 2021). During the last glacial maximum, the sea level was up to 130 m lower than present, and the seamounts of the Vitoria-Trindade chain (VTC) were above the surface and closer to each other, therefore favoring the dispersal of organisms that use shallow habitats (Lambeck et al. 2002). The colonization of TRI by S. axillare likely occurred during a glacial period, when conditions for dispersal using the seamounts as stepping stones were more favorable. When the sea level increased, local extinction of S. axillare is likely to have occurred on the seamounts, explaining why the species is not recorded on the VTC (Pinheiro et al. 2015). The environmental variability during glacial cycles also affected other species in the region. Analyses of endemic species from VTC and TRI and its mainland sister species indicate intermittent connectivity and colonization events following growth of the mainland population (Pinheiro et al. 2017). For reef species with a higher capacity for colonization, the seamounts of VTC are currently functioning as stepping stones and contribute to genetic connectivity (Thomas et al. 2009; Pinheiro et al. 2009; Simon et al. 2013, 2021; Macieira et al. 2015). However, for weak colonizers, and those species that are restricted to shallower waters, low gene flow results in genetic differentiation (Simon et al. 2021). Consequently, most of the VTC and TRI endemic lineages are weak colonizers (Pinheiro et al. 2017).
Although S. axillare has never been reported on the VTC seamounts, other species of Sparisoma have, including its closest relative, S. frondosum, that is present on the seamounts closer to the continent and absent in TRI (Pinheiro et al. 2015). Because of the close phylogenetic relationships, similarities in life-history attributes and distribution patterns, it was suggested that S. axillare and S. frondosum have a similar colonization capacity and habitat preference (Peterson et al. 1999; Losos 2011). However, it has been subsequently shown that S. axillare has different habitat requirements to S. frondosum and this has been attributed to divergent evolution of ‘sister species’ to avoid competition (Schoener 1974; Dufour et al. 2015; Pinheiro et al. 2015). Our data suggest that these species also have different capacities for colonization, further supported by evidence for long-distance dispersal of S. frondosum across deep ocean to recently colonize Cape Verde (Freitas et al. 2014). Similar comparisons within the region of our study for the reef fishes Acanthurus (Rocha et al. 2002) and Halichoeres (Rocha et al. 2005) indicated divergent habitat preferences among sister species in terms of depth, temperature and bottom type. Collectively, these observations argue against the proposition that knowledge of genetic structure for one species can be used as a proxy for genetic structure in closely related species.
The contrasting levels of genetic isolation of S. axillare at the off-coast sites of FNA and TRI can be explained by different oceanographic conditions, principally the currents and geographic distance (Lumpkin and Garzoli 2005; Rodrigues et al. 2007; Rudorff et al. 2009). FNA is affected by the Central South Equatorial Current and the North Brazil Current, which includes strong currents that flow from the east toward the coast. TRI, which is located more than 1000 km off the coast, is dominated by the weaker Brazil Current, which flows from the north to the south (Stramma and England 1999) and by eddies that mostly affect the VTC. Some genetic studies in the same area on fish and corals contrast with our results by showing significant genetic divergence between coast and FNA, with FNA occasionally clustering together with TRI, despite the almost 2 thousand kilometers of distance across open ocean (Neves et al. 2016; Peluso et al. 2018). Nonetheless, our results flag the importance of FNA in species conservation because of its possible role as a source population (Endo et al. 2019).
Consistent with previous biophysical models for Sparisoma larvae, low genetic divergence was evident within the Costa dos Corais MPA at PE and AL (Endo et al. 2019), an important region embracing a large diversity of Brazilian reef fauna (Pereira et al. 2021). These data are consistent with the idea that this region is key to maintaining populations sizes and genetic variation of S. axillare (Endo et al. 2019). The importance of the Costa dos Corais MPA for the recruitment of S. axillare and, probably, other shallow water reef species, emphasizes the need to ensure sustainable levels of exploitation in this MPA as recommended by an IUCN category IV (Dudley 2008). In this MPA, the only enforced management measures in place are small coastal no-take zones (ICMBIO 2020). The other regions are open for fishing, possibly leading to a reduction of the local S. axillare stock, thereby threatening the species along its entire coastal distribution, especially if the surrounding areas are also strongly exploited. The lack of protection is further highlighted by ABR, which is the largest coralline bank in the south Atlantic and only 1% of its total area is fully protected. This site also showed some evidence for S. axillare being genetic differentiated from other parts of its distribution, suggesting that this region may be less likely replenished by migrants and less likely to act as a source to other parts of the distribution.
Knowledge of connectivity is crucial to conservation planning, and, in the last decade, genetic and habitat connectivity measures have been adopted as criteria in integrative management initiatives (Saura et al. 2019). Due to high levels of commercial, artisanal and recreational fishing within its distribution along the Brazilian coast, S. axillare has the status of vulnerable (Roos et al. 2015, 2016; MMA 2022). However, this species is only effectively protected in a few MPAs along the entire Brazilian Province (Morais et al. 2017; Endo et al. 2019; Giglio et al. 2018). Here we show that parts of the distribution of S. axillare are genetically divergent, despite its comparative long pelagic larval phase. The S. axillare population at TRI could be especially vulnerable to the effects of overfishing given the level genetic divergence and inferred isolation, and because the likely direction of any migration is toward the coast (Pinheiro et al. 2010). Although at TRI and other oceanic islands (São Pedro and São Paulo Archipelago) no-take areas have recently been established, thus increasing the country’s marine protected areas, these MPAs are still loosely regulated to commercial and recreational fisheries (Guabiroba et al. 2020). Our results support the notion that the Brazilian oceanic islands are an important repository of unique genetic variation (Pinheiro et al. 2015, 2017; Mazzei et al. 2021).
References
Albaina A, Iriondo M, Velado I, Laconcha U, Zarraonaindia I, Arrizabalaga H, Pardo MA, Lutcavage M, Grant WS, Estonba A (2013) Single nucleotide polymorphism discovery in albacore and Atlantic bluefin tuna provides insights into worldwide population structure. Anim Genet 44(6):678–692. https://doi.org/10.1111/age.12051
Assis J, Tyberghein L, Bosch S, Verbruggen H, Serrão EA, De Clerck O. Bio-ORACLE v2. 0: extending marine data layers for bioclimatic modelling. Glob Ecol Biogeogr 27(3):277–284. https://doi.org/10.1111/geb.12693
Baggio RA, Stoiev SB, Spach HL, Boeger WA (2017) Opportunity and taxon pulse: the central influence of coastal geomorphology on genetic diversification and endemism of strict estuarine species. J Biogeogr 44:1626–1639. https://doi.org/10.1111/jbi.12934
Baker P, Austin JD, Bowen BW, Baker SM (2008) Range-wide population structure and history of the northern quahog (Merceneria merceneria) inferred from mitochondrial DNA sequence data. ICES J Mar Sci 65(2):155–163. https://doi.org/10.1093/icesjms/fsn007
Barbosa MC, Luiz OJ, Cordeiro CAMM, Giglio VJ, Ferreira CEL (2021) Fish and spearfisher traits contributing to catch composition. Fish Res 241:105988. https://doi.org/10.1016/j.fishres.2021.105988
Beheregaray LB, Sunnucks P, Briscoe DA (2002) A rapid fish radiation associated with the last sea-level changes in southern Brazil: the silverside Odontesthes perugiae complex. Proc R Soc B: Biol Sci 269:65–73. https://doi.org/10.1098/rspb.2001.1838
Bender MG, Machado GR, de Azevedo Silva PJ, Floeter SR, Monteiro-Netto C, Luiz OJ, Ferreira CEL (2014) Local ecological knowledge and scientific data reveal overexploitation by multigear artisanal fisheries in the Southwestern Atlantic. PLOS ONE 9:e110332. https://doi.org/10.1371/journal.pone.0110332
Bowen BW, Muss A, Rocha LA, Grant WS (2006) Shallow mtDNA coalescence in Atlantic Pygmy Angelfishes (Genus Centropyge) indicates a recent invasion from the Indian Ocean. J Hered 97:1–12. https://doi.org/10.1093/jhered/esj006
Bradbury IR, Hubert S, Higgins B, Bowman S, Borza T, Paterson IG, Snelgrove PV, Morris CJ, Gregory RS, Hardie D, Hutchings JA (2013) Genomic islands of divergence and their consequences for the resolution of spatial structure in an exploited marine fish. Evol Appl 6(3):450–461. https://doi.org/10.1111/eva.12026
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. JABES 7(3):361–372. https://doi.org/10.1198/108571102320
Clements KD, German DP, Piché J, Tribollet A, Choat JH (2017) Integrating ecological roles and trophic diversification on coral reefs: multiple lines of evidence identify parrotfishes as microphages. Biol J Linn Soc 120:729–751. https://doi.org/10.1111/bij.12914
Córdova-Alarcón VR, Araneda C, Jilberto F, Magnolfi P, Toledo MI, Lam N (2019) Genetic diversity and population structure of Genypterus chilensis, a commercial benthic marine species of the South Pacific. Front Mar Sci 12(6):748. https://doi.org/10.3389/fmars.2019.00748
Cunningham KM, Canino MF, Spies IB, Hauser L (2009) Genetic isolation by distance and localized fjord population structure in Pacific cod (Gadus macrocephalus): limited effective dispersal in the northeastern Pacific Ocean. Can J Fish Aquat Sci 66(1):153–166. https://doi.org/10.1139/F08-199
da Silva WC, Marceniuk AP, Sales JBL, Araripe J (2016) Early Pleistocene lineages of Bagre bagre (Linnaeus, 1766) (Siluriformes: Ariidae), from the Atlantic coast of South America, with insights into the demography and biogeography of the species. Neotrop Ichthyol. https://doi.org/10.1590/1982-0224-20150184
Dias RM, Lima SMQ, Mendes LF, Almeida DF, Paiva PC, Britto MR (2019) Different speciation processes in a cryptobenthic reef fish from the Western Tropical Atlantic. Hydrobiologia 837:133–147. https://doi.org/10.1007/s10750-019-3966-z
Dixo M, Metzger JP, Morgante JS, Zamudio KR (2009) Habitat fragmentation reduces genetic diversity and connectivity among toad populations in the Brazilian Atlantic Coastal Forest. Biol Conserv 142:1560–1569. https://doi.org/10.1016/j.biocon.2008.11.016
Dray S, Dufour A-B (2007) The ade4 Package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20. https://doi.org/10.18637/jss.v022.i04
Dudley N (2008) Guidelines for applying protected area management categories. IUCN
Dufour CMS, Meynard C, Watson J, Rioux C, Benhamou S, Perez J, du Plessis JJ, Avenant N, Pillay N, Ganem G (2015) Space use variation in co-occurring sister species: response to environmental variation or competition? PLoS ONE 10:e0117750. https://doi.org/10.1371/journal.pone.0117750
Eggertsen L, Ferreira CEL, Fontoura L, Kautsky N, Gullström M, Berkström C (2017) Seaweed beds support more juvenile reef fish than seagrass beds in a south-western Atlantic tropical seascape. Estuar Coast Shelf Sci 196:97–108. https://doi.org/10.1016/j.ecss.2017.06.041
Endo CAK, Gherardi DFM, Pezzi LP, Lima LN (2019) Low connectivity compromises the conservation of reef fishes by marine protected areas in the tropical South Atlantic. Sci Rep 9:8634. https://doi.org/10.1038/s41598-019-45042-0
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Feitosa JLL, Ferreira BP (2015) Distribution and feeding patterns of juvenile parrotfish on algal-dominated coral reefs. Mar Ecol 36:462–474. https://doi.org/10.1111/maec.12154
Ferreira CEL, Gonçalves JEA (2006) Community structure and diet of roving herbivorous reef fishes in the Abrolhos Archipelago, south-western Atlantic. J Fish Biol 69:1533–1551. https://doi.org/10.1111/j.1095-8649.2006.01220.x
Ferreira CEL, Floeter SR, Gasparini JL, Ferreira BP, Joyeux JC (2004) Trophic structure patterns of Brazilian reef fishes: a latitudinal comparison. J Biogeogr 31:1093–1106. https://doi.org/10.1111/j.1365-2699.2004.01044.x
Floeter SR, Rocha LA, Robertson DR, Joyeux JC, Smith-Vaniz WF, Wirtz P, Edwards AJ, Barreiros JP, Ferreira CEL, Gasparini JL, Brito A, Falcón JM, Bowen BW, Bernardi G (2008) Atlantic reef fish biogeography and evolution. J Biogeogr 35:22–47. https://doi.org/10.1111/j.1365-2699.2007.01790.x
Freitas R, Luiz OJ, Silva PN, Floeter SR, Bernardi G, Ferreira CEL (2014) The occurrence of Sparisoma frondosum (Teleostei: Labridae) in the Cape Verde Archipelago, with a summary of expatriated Brazilian endemic reef fishes. Mar Biodiv 44:173–179. https://doi.org/10.1007/s12526-013-0194-z
Giglio VJ, Pinheiro HT, Bender MG, Bonaldo RM, Costa-Lotufo LV, Ferreira CEL, Floeter SR, Freire A, Gasparini JL, Joyeux J-C, Krajewski JP, Lindner A, Longo GO, Lotufo TMC, Loyola R, Luiz OJ, Macieira RM, Magris RA, Mello TJ, Quimbayo JP, Rocha LA, Segal B, Teixeira JB, Vila-Nova DA, Vilar CC, Zilberberg C, Francini-Filho RB (2018) Large and remote marine protected areas in the South Atlantic Ocean are flawed and raise concerns: comments on Soares and Lucas (2018). Mar Pol 96:13–17. https://doi.org/10.1016/j.marpol.2018.07.017
Gilg MR, Hilbish TJ (2003) The geography of marine larval dispersal: coupling genetics with fine-scale physical oceanography. Ecology 84:2989–2998. https://doi.org/10.1890/02-0498
Gruber B, Unmack PJ, Berry OF, Georges A (2018) Dartr: an r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Res 18:691–699. https://doi.org/10.1111/1755-0998.12745
Guabiroba HC, Santos MEA, Pinheiro HT, Simon T, Pimentel CR, Vilar CC, Joyeux J-C (2020) Trends in recreational fisheries and reef fish community structure indicate decline in target species population in an isolated tropical oceanic island. Ocean Coast Manag 191:105194. https://doi.org/10.1016/j.ocecoaman.2020.105194
Guo X, Zhao D, Jung D, Li Q, Kong L-F, Ni G, Nakano T, Matsukuma A, Kim S, Park C, Lee HJ, Park J-K (2015) Phylogeography of the Rock Shell Thais clavigera (Mollusca): evidence for long-distance dispersal in the Northwestern Pacific. PLoS ONE 10:e0129715. https://doi.org/10.1371/journal.pone.0129715
ICMBIO (2020) Plano de manejo da Área de Proteção Ambiental Costa dos Corais. https://www.icmbio.gov.br/apacostadoscorais/planos-de-manejo/zoneamento.html (accessed December 15, 2021)
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity Arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res 29:e25. https://doi.org/10.1093/nar/29.4.e25
Johnson MS, Black R (2006) Islands increase genetic subdivision and disrupt patterns of connectivity of intertidal snails in a complex archipelago. Evolution 60:2498–2506. https://doi.org/10.1111/j.0014-3820.2006.tb01885.x
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Lambeck K, Esat TM, Potter E-K (2002) Links between climate and sea levels for the past three million years. Nature 419:199–206. https://doi.org/10.1038/nature01089
Liedke A, Pinheiro H, Floeter S, Bernardi G (2020) Phylogeography of the banded butterflyfish, Chaetodon striatus, indicates high connectivity between biogeographic provinces and ecosystems in the western Atlantic. Neotrop Ichthyol. https://doi.org/10.1590/1982-0224-2019-0054
Longo GO, Hay ME, Ferreira CEL, Floeter SR (2019) Trophic interactions across 61 degrees of latitude in the Western Atlantic. Glob Ecol Biogeogr 28:107–117. https://doi.org/10.1111/geb.12806
Losos JB (2011) Convergence, adaptation, and constraint. Evolution 65:1827–1840. https://doi.org/10.1111/j.1558-5646.2011.01289.x
Ludt WB, Rocha LA (2015) Shifting seas: the impacts of Pleistocene sea-level fluctuations on the evolution of tropical marine taxa. J Biogeogr 42:25–38. https://doi.org/10.1111/jbi.12416
Luiz OJ, Madin JS, Robertson DR, Rocha LA, Wirtz P, Floeter SR (2012) Ecological traits influencing range expansion across large oceanic dispersal barriers: insights from tropical Atlantic reef fishes. Proc R Soc B: Biol Sci 279:1033–1040. https://doi.org/10.1098/rspb.2011.1525
Luiz OJ, Allen AP, Robertson DR, Floeter SR, Kulbicki M, Vigliola L, Becheler R, Madin JS (2013) Adult and larval traits as determinants of geographic range size among tropical reef fishes. PNAS 110:16498–16502. https://doi.org/10.1073/pnas.1304074110
Lumpkin R, Garzoli SL (2005) Near-surface circulation in the Tropical Atlantic Ocean. Deep Sea Res Part I: Oceanogr Res Pap 52:495–518. https://doi.org/10.1016/j.dsr.2004.09.001
Lundy CJ, Moran P, Rico C, Milner RS, Hewitt GM (1999) Macrogeographical population differentiation in oceanic environments: a case study of European hake (Merluccius merluccius), a commercially important fish. Mol Ecol 8(11):1889–1898. https://doi.org/10.1046/j.1365-294x.1999.00789.x
Machado LF, de Souza Damasceno J, Bertoncini ÁA, Tosta VC, Farro APC, Hostim-Silva M, Oliveira C (2017) Population genetic structure and demographic history of the spadefish, Chaetodipterus faber (Ephippidae) from Southwestern Atlantic. J Exp Mar Biol Ecol 487:45–52. https://doi.org/10.1016/j.jembe.2016.11.005
Macieira RM, Simon T, Pimentel CR, Joyeux J-C (2015) Isolation and speciation of tidepool fishes as a consequence of Quaternary sea-level fluctuations. Environ Biol Fish 98:385–393. https://doi.org/10.1007/s10641-014-0269-0
Marceniuk AP, Burlamaqui TCT, Oliveira C, Carneiro J, Soares BE, de Luna Sales JB (2019) Incipient speciation, driven by distinct environmental conditions, in the marine catfishes of the genus Aspistor (Siluriformes, Ariidae), from the Atlantic coast of South America. J Zool Syst Evol Res 57:400–417. https://doi.org/10.1111/jzs.12261
Mazzei EF, Pinheiro HT, Morais RA, Floeter SR, Veras DP, Queiroz LV, Joyeux J-C, Ferreira CEL (2019) Parrotfishes of the genus Scarus in southwestern Atlantic oceanic reef environments: occasional pulse or initial colonization? Mar Biodiv 49:555–561. https://doi.org/10.1007/s12526-017-0827-8
Mazzei EF, Pinheiro HT, Simon T, Moura RL, Macieira RM, Pimentel CR, Teixeira JB, Floeter SR, Ferreira CEL, Ghisolfi RD, Francini-Filho RB, Quimbayo JP, Rocha LA, Gasparini JL, Joyeux J-C (2021) Mechanisms of dispersal and establishment drive a stepping stone community assembly on seamounts and oceanic islands. Mar Biol 168:109. https://doi.org/10.1007/s00227-021-03919-7
McRae BH, Beier P (2007) Circuit theory predicts gene flow in plant and animal populations. PNAS 104:19885–19890. https://doi.org/10.1073/pnas.0706568104
MMA (2022) Brazilian List of Endangered Aquatic Animals
Mora C, Sale PF (2002) Are populations of coral reef fish open or closed? TREE 17:422–428. https://doi.org/10.1016/S0169-5347(02)02584-3
Morais RA, Ferreira CEL, Floeter SR (2017) Spatial patterns of fish standing biomass across Brazilian reefs. J Fish Biol 91:1642–1667. https://doi.org/10.1111/jfb.13482
Neves JMM, Lima SMQ, Mendes LF, Torres RA, Pereira RJ, Mott T (2016) Population structure of the Rockpool Blenny Entomacrodus vomerinus shows source-sink dynamics among ecoregions in the Tropical Southwestern Atlantic. PLoS ONE 11:e0157472. https://doi.org/10.1371/journal.pone.0157472
Nunes FLD, Norris RD, Knowlton N (2011) Long distance dispersal and connectivity in amphi-atlantic corals at regional and basin scales. PLoS ONE 6:e22298. https://doi.org/10.1371/journal.pone.0022298
Peluso L, Tascheri V, Nunes FLD, Castro CB, Pires DO, Zilberberg C (2018) Contemporary and historical oceanographic processes explain genetic connectivity in a Southwestern Atlantic coral. Sci Rep 8:2684. https://doi.org/10.1038/s41598-018-21010-y
Pereira PHC, Côrtes LGF, Lima GV, Gomes E, Pontes AVF, Mattos F, Araújo ME, Ferreira-Junior F, Sampaio CLS (2021) Reef fishes biodiversity and conservation at the largest Brazilian coastal Marine Protected Area (MPA Costa dos Corais). Neotrop Ichthyol 19:e210071. https://doi.org/10.1590/1982-0224-2021-0071
Peterman WE (2018) ResistanceGA: an R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol Evol 9:1638–1647. https://doi.org/10.1111/2041-210X.12984
Peterson AT, Soberón J, Sánchez-Cordero V (1999) Conservatism of ecological niches in evolutionary time. Science. https://doi.org/10.1126/science.285.5431.126
Pinheiro H, Camilato V, Luiz J, Joyeux J-C (2009) New records of fishes for Trindade-Martin Vaz oceanic insular complex, Brazil. Zootaxa 2298
Pinheiro HT, Martins AS, Gasparini JL (2010) Impact of commercial fishing on Trindade Island and Martin Vaz Archipelago, Brazil: characteristics, conservation status of the species involved and prospects for preservation. Braz Arch Biol Technol 53:1417–1423. https://doi.org/10.1590/S1516-89132010000600018
Pinheiro HT, Mazzei E, Moura RL, Amado-Filho GM, Carvalho-Filho A, Braga AC, Costa PAS, Ferreira BP, Ferreira CEL, Floeter SR, Francini-Filho RB, Gasparini JL, Macieira RM, Martins AS, Olavo G, Pimentel CR, Rocha LA, Sazima I, Simon T, Teixeira JB, Xavier LB, Joyeux J-C (2015) Fish biodiversity of the Vitória-Trindade Seamount Chain, Southwestern Atlantic: an updated database. PLoS ONE 10:e0118180. https://doi.org/10.1371/journal.pone.0118180
Pinheiro HT, Bernardi G, Simon T, Joyeux J-C, Macieira RM, Gasparini JL, Rocha C, Rocha LA (2017) Island biogeography of marine organisms. Nature 549:82–85. https://doi.org/10.1038/nature23680
Pinheiro HT, Rocha LA, Macieira RM, Carvalho-Filho A, Anderson AB, Bender MG, Di Dario F, Ferreira CEL, Figueiredo-Filho J, Francini-Filho R, Gasparini JL, Joyeux J-C, Luiz OJ, Mincarone MM, Moura RL, de Anchieta C. C. Nunes J, Quimbayo JP, Rosa RS, Sampaio CLS, Sazima I, Simon T, Vila-Nova DA, Floeter SR (2018) South-western Atlantic reef fishes: zoogeographical patterns and ecological drivers reveal a secondary biodiversity centre in the Atlantic Ocean. Divers Distrib 24:951–965. https://doi.org/10.1111/ddi.12729
Pinheiro HT, Nunes JACC, Coni EOC, Almeida ECG, Sampaio CLS, Ferreira CEL, Meirelles PM, Hostim-Silva M, Pereira PHC, Giglio VJ, Gasparini JL, Rocha LA, Ferreira CM (2021) An inverted management strategy for the fishery of endangered marine species. Front Mar Sci 8:172. https://doi.org/10.3389/fmars.2021.604108
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959. https://doi.org/10.1093/genetics/155.2.945
R Core Team (2019) R: a language and environment for statistical computing. https://www.r-project.org/ (accessed December 15, 2021)
Riginos C, Nachman MW (2001) Population subdivision in marine environments: the contributions of biogeography, geographical distance and discontinuous habitat to genetic differentiation in a blennioid fish, Axoclinus nigricaudus. Mol Ecol 10:1439–1453. https://doi.org/10.1046/j.1365-294X.2001.01294.x
Robertson DR, Karg F, Leão de Moura R, Victor BC, Bernardi G (2006) Mechanisms of speciation and faunal enrichment in Atlantic parrotfishes. Mol Phylogen Evol 40:795–807. https://doi.org/10.1016/j.ympev.2006.04.011
Rocha LA (2003) Patterns of distribution and processes of speciation in Brazilian reef fishes. J Biogeogr 30:1161–1171. https://doi.org/10.1046/j.1365-2699.2003.00900.x
Rocha LA, Bass AL, Robertson DR, Bowen BW (2002) Adult habitat preferences, larval dispersal, and the comparative phylogeography of three Atlantic surgeonfishes (Teleostei: Acanthuridae). Mol Ecol 11:243–251. https://doi.org/10.1046/j.0962-1083.2001.01431.x
Rocha LA, Robertson DR, Roman J, Bowen BW (2005) Ecological speciation in tropical reef fishes. Proc R Soc B: Biol Sci 272:573–579. https://doi.org/10.1098/2004.3005
Rocha LA, Craig MT, Bowen BW (2007) Phylogeography and the conservation of coral reef fishes. Coral Reefs 26:501–512. https://doi.org/10.1007/s00338-007-0261-7
Rodrigues RR, Rothstein LM, Wimbush M (2007) Seasonal variability of the South Equatorial Current bifurcation in the Atlantic Ocean: a numerical study. J Phys Oceanogr 37:16–30. https://doi.org/10.1175/JPO2983.1
Roos NC, Carvalho AR, Lopes PFM, Pennino MG (2015) Modeling sensitive parrotfish (Labridae: Scarini) habitats along the Brazilian coast. Mar Environ Res 110:92–100. https://doi.org/10.1016/j.marenvres.2015.08.005
Roos NC, Pennino MG, de Macedo Lopes PF, Carvalho AR (2016) Multiple management strategies to control selectivity on parrotfishes harvesting. Ocean Coast Manag 134:20–29. https://doi.org/10.1016/j.ocecoaman.2016.09.029
Roos NC, Pennino MG, Carvalho AR, Longo GO (2019) Drivers of abundance and biomass of Brazilian parrotfishes. Mar Ecol Prog Ser 623:117–130. https://doi.org/10.3354/meps13005
Roy EM, Quattro JM, Greig TW (2012) Genetic management of Black Sea Bass: influence of biogeographic barriers on population structure. Mar Coast Fish 4(1):391–402. https://doi.org/10.1080/19425120.2012.675983
Rudorff CAG, Lorenzzetti JA, Gherardi DFM, Lins-Oliveira JE (2009) Application of remote sensing to the study of the pelagic spiny lobster larval transport in the Tropical Atlantic. Braz J Oceanogr 57:7–16. https://doi.org/10.1590/S1679-87592009000100002
Saenz-Agudelo P, Dibattista JD, Piatek MJ, Gaither MR, Harrison HB, Nanninga GB, Berumen ML (2015) Seascape genetics along environmental gradients in the Arabian Peninsula: insights from ddRAD sequencing of anemonefishes. Mol Ecol 24:6241–6255. https://doi.org/10.1111/mec.13471
Sánchez P, Alvarez JA (1988) Scaeurgus unicirrhus (Orbigny, 1840) (Cephalopoda, Octopodidae): first record from the South-East Atlantic. S Afr J Mar Sci 7:69–74
Santos S, Hrbek T, Farias IP, Schneider H, Sampaio I (2006) Population genetic structuring of the king weakfish, Macrodon ancylodon (Sciaenidae), in Atlantic coastal waters of South America: deep genetic divergence without morphological change. Mol Ecol 15:4361–4373. https://doi.org/10.1111/j.1365-294X.2006.03108.x
Saura S, Bertzky B, Bastin L, Battistella L, Mandrici A, Dubois G (2019) Global trends in protected area connectivity from 2010 to 2018. Biol Conserv 238:108183. https://doi.org/10.1016/j.biocon.2019.07.028
Sbrocco EJ, Barber PH (2013) MARSPEC: ocean climate layers for marine spatial ecology. Ecology 94:979–979. https://doi.org/10.1890/12-1358.1
Schoener TW (1974) Resource Partitioning in Ecological Communities. Science
Simon T, Macieira RM, Joyeux J-C (2013) The shore fishes of the Trindade–Martin Vaz insular complex: an update. J Fish Biol 82:2113–2127. https://doi.org/10.1111/jfb.12126
Simon T, Pinheiro HT, Santos S, Macieira RM, Ferreira YSS, Bernardi G, Rocha LA, Floeter SR, Ferreira CEL, Joyeux J-C (2021) Comparative phylogeography of reef fishes indicates seamounts as stepping stones for dispersal and diversification. Coral Reefs. https://doi.org/10.1007/s00338-021-02178-8
Siqueira AC, Bellwood DR, Cowman PF (2019) The evolution of traits and functions in herbivorous coral reef fishes through space and time. Proc R Soc B: Biol Sci 286:20182672. https://doi.org/10.1098/rspb.2018.2672
Stramma L, England M (1999) On the water masses and mean circulation of the South Atlantic Ocean. J Geophys Res: Oceans 104:20863–20883. https://doi.org/10.1029/1999JC900139
Strugnell JM, Allcock AL, Watts PC (2017) Closely related octopus species show different spatial genetic structures in response to the Antarctic seascape. Ecol Evol 7:8087–8099. https://doi.org/10.1002/ece3.3327
Takeuchi T, Masaoka T, Aoki H, Koyanagi R, Fujie M, Satoh N (2020) Divergent northern and southern populations and demographic history of the pearl oyster in the western Pacific revealed with genomic SNPs. Evol Appl 13:837–853. https://doi.org/10.1111/eva.12905
Teschima M, Ströher P, Strӧher R, Firkowski C, Pie M, Freire A (2016) Large-scale connectivity of Grapsus grapsus (Decapoda) in the Southwestern Atlantic oceanic islands: integrating genetic and morphometric data. Mar Ecol 37:1360–1372. https://doi.org/10.1111/maec.12347
Thomas AL, Henderson GM, Deschamps P, Yokoyama Y, Mason AJ, Bard E, Hamelin B, Durand N, Camoin G (2009) Penultimate deglacial sea-level timing from uranium/thorium dating of tahitian corals. Science 324:1186–1189. https://doi.org/10.1126/science.1168754
Treml EA, Halpin PN, Urban DL, Pratson LF (2008) Modeling population connectivity by ocean currents, a graph-theoretic approach for marine conservation. Landscape Ecol 23:19–36. https://doi.org/10.1007/s10980-007-9138-y
Truelove NK, Kough AS, Behringer DC, Paris CB, Box SJ, Preziosi RF, Butler MJ (2017) Biophysical connectivity explains population genetic structure in a highly dispersive marine species. Coral Reefs 36(1):233–244. https://doi.org/10.1007/s00338-016-1516-y
Tyberghein L, Verbruggen H, Pauly K, Troupin C, Mineur F, De Clerck O (2012) Bio-ORACLE: a global environmental dataset for marine species distribution modelling. Glob Ecol Biogeogr 21:272–281. https://doi.org/10.1111/j.1466-8238.2011.00656.x
Victor BC, Wellington GM (2000) Endemism and the pelagic larval duration of reef fishes in the eastern Pacific Ocean. Mar Ecol Prog Ser 205:241–248. https://doi.org/10.3354/meps205241
Volk DR, Konvalina JD, Floeter SR, Ferreira CEL, Hoffman EA (2021) Going against the flow: barriers to gene flow impact patterns of connectivity in cryptic coral reef gobies throughout the western Atlantic. J Biogeogr 48:427–439. https://doi.org/10.1111/jbi.14010
Weersing K, Toonen RJ (2009) Population genetics, larval dispersal, and connectivity in marine systems. Mar Ecol Prog Ser 393:1–12. https://doi.org/10.3354/meps08287
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001 and part by MCTI/CNPq/Universal—424790/2016-5 and by National Geographic Society—Research and Exploration (CP-077ER-17). JTV was funded by a PhD scholarship—CAPES and Macquarie University PhD scholarship. BPF (309216/2018-4). PFML (301515/2019-0) and SMQL (313644/2018-7) thank CNPq for productivity grants. CELF is funded by FAPERJ and CNPq. We thank R. Ranulpho for the map figure and the UFRN Graduate Program in Ecology for the infrastructural support.
Funding
Open Access funding enabled and organized by Projekt DEAL. This study was supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Finance Code 001, the National Geographic Society, the Conselho Nacional de Desenvolvimento Científico e Tecnológico—Brasil (CNPq) Edital Universal and by National Geographic Society—Research and Exploration (CP-077ER-17).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest. Most samples were collected from dead animals in fish markets. The permit for data collection in situ was approved by SISBIO (authorization number 48112-7).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tovar Verba, J., Ferreira, C.E.L., Pennino, M.G. et al. Genetic structure of the threatened Gray Parrotfish (Sparisoma axillare) in the Southwestern Atlantic. Coral Reefs 42, 105–117 (2023). https://doi.org/10.1007/s00338-022-02324-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-022-02324-w