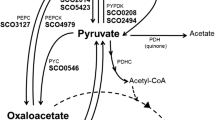
Abstract
In Kluyveromyces lactis, the casein kinase I (Rag8p) regulates the transcription of glycolytic genes and the expression of the low-affinity glucose transporter gene RAG1. This control involves the transcription factor Sck1p, a homologue of Sgc1p of Saccharomyces cerevisiae. SGC1 is known to interact genetically with ScGCR1 and ScGCR2, which code for regulators of glycolytic gene expression. Therefore, we studied the role of KlGCR1 and KlGCR2 genes in K. lactis. The Klgcr1 null mutant could not grow on glucose when respiration was blocked by antimycin A (Rag− phenotype). In contrast, the Klgcr2 null mutant could grow under the same conditions, although at a reduced rate. In both mutants, the transcription of glycolytic genes was affected, while that of ribosomal protein genes was not modified. Furthermore, the transcription of the glucose permease genes was also found to be affected in the two mutants, although dissimilarly. While RAG1 transcription decreased at high glucose concentrations, the expression of the high-affinity glucose permease gene HGT1 was unexpectedly impaired under gluconeogenic conditions, in the absence of glucose. Gel mobility shift assays performed with purified maltose-binding protein–KlGcr1p showed that KlGcr1p could interact directly with the promoters of the glycolytic genes, but not with the promoters of the glucose permease genes. Thus, the control exerted by KlGcr1p and KlGcr2p upon glucose transporter genes is probably indirect.








Similar content being viewed by others
References
Baker HV (1991) GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc Natl Acad Sci USA 88:9443–9447
Betina S, Goffrini P, Ferrero I, Wésolowski-Louvel M (2001) RAG4 gene encodes a glucose sensor in Kluyveromyces lactis. Genetics 158:541–548
Bianchi MM, Tizzani L, Destruelle M, Frontali L, Wésolowski-Louvel M (1996) The “petite-negative” yeast Kluyveromyces lactis has a single pyruvate decarboxylase gene. Mol Microbiol 19:22–36
Billard P, Ménart S, Blaisonneau J, Bolotin-Fukuhara M, Fukuhara H, Wésolowski-Louvel M (1996) Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J Bacteriol 178:5860–5866
Blaisonneau J, Fukuhara H, Wésolowski-Louvel M (1997) The Kluyveromyces lactis equivalent of casein kinase I is required for the transcription of the gene encoding the low-affinity glucose permease. Mol Gen Genet 253:469–477
Boles E, Zimmermann FK (1993) Induction of pyruvate decarboxylase in glycolysis mutants of Saccharomyces cerevisiae correlates with the concentrations of three-carbon glycolytic metabolites. Arch Microbiol 160:324–328
Bolotin-Fukuhara M, Toffano-Nioche C, Artiguenave F, Duchateau-Nguyen G, Lemaire M, Marmeisse R, Montrocher R, Robert C, Termier M, Wincker P, Wésolowski-Louvel M (2000) Genomic exploration of the hemiascomycetous yeasts: 11. Kluyveromyces lactis. FEBS Lett 487:66–70
Breunig KD, Bolotin-Fukuhara M, Bianchi MM, Bourgarel D, Falcone C, Ferrero I, Frontali L, Goffrini P, Kruger JJ, Mazzoni C, Milkowski C, Steensma Y, Wésolowski-Louvel M, Zeeman AM (2000) Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb Technol 26:771–780
Chambers A, Packham EA, Graham IR (1995) Control of glycolytic gene expression in the budding yeast (Saccharomyces cerevisiae) Curr Genet 29:1–9
Chen XJ (1996) Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131–136
Chen XJ, Wésolowski-Louvel M, Fukuhara H (1992) Glucose transport in the yeast Kluyveromyces lactis. II. Transcriptional regulation of the glucose transporter gene RAG1. Mol Gen Genet 33:97–105
Clifton D, Fraenkel DG (1981) The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem 256:13074–13078
Deminoff SJ, Santangelo GM (2001) Rap1p requires Gcr1p and Gcr2p homodimers to activate ribosomal protein and glycolytic genes, respectively. Genetics 158:133–143
Dohmen RJ, Strasser AWM, Höner CB, Hollenberg CP (1991) An efficient transformation procedure enabling long-term storage of competent cells of various yeast genera. Yeast 7:691–692
Drazinic CM, Smerage JB, Lopez MC, Baker HV (1996) Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p) Mol Cell Biol 16:3187–3196
Gietz RD, Sugino A (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534
Goffrini P, Algeri AA, Donnini C, Wésolowski-Louvel M, Ferrero I (1989) RAG1 and RAG2: nuclear genes involved in the dependence/independence on mitochondrial respiratory function for the growth on sugars. Yeast 5:99–106
Goffrini P, Wésolowski-Louvel M, Ferrero I (1991) A phosphoglucose isomerase gene is involved in the Rag phenotype of the yeast Kluyveromyces lactis. Mol Gen Genet 228:401–409
Goncalves PM, Griffoen G, Hbelman JP, Planta RJ (1997) Signaling pathways leading to transcriptional regulation of genes involved in the activation of glycolysis in yeast. Mol Microbiol 25:483–493
Haw R, Yarragudi AD, Uemura H (2001) Isolation of GCR1, a major transcription factor of glycolytic genes in Saccharomyces cerevisiae, from Kluyveromyces lactis. Yeast 18:729–735
Holland MJ, Yokoi T, Holland JP, Myambo K, Innis MA (1987) The GCR1 gene encodes a positive transcriptional regulator of the enolase and glyceraldehyde-3-phosphate dehydrogenase gene families in Saccharomyces cerevisiae. Mol Cell Biol 7:813–820
Huie MA, Scott EW, Drazinic CM, Lopez MC, Hornstra IK, Yang TP, Baker HV (1992) Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol 12:2690–2700
Iost I, Dreyfus M (1995) The stability of Escherichia coli lacZ mRNA depends upon the simultaneity of its synthesis and translation. EMBO J 14:3252–3561
Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153:163–168
Kief DR, Warner JR (1981) Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol 1:1007–1015
Larson PG, Rossi JJ (1991) Altered response to growth rate changes in Kluyveromyces lactis versus Saccharomyces cerevisiae as demonstrated by heterologous expression of ribosomal protein 59 (CRY1). Nucleic Acids Res 19:4701–4707
Lemaire M, Guyon A, Betina S, Wésolowski-Louvel M (2002) Regulation of glycolysis by casein kinase I (Rag8p) in Kluyveromyces lactis involves a DNA-binding protein, Sck1p, a homologue of Sgc1p of Saccharomyces cerevisiae. Curr Genet 40:355–364
Lopez MC, Baker HV (2000) Understanding the growth phenotype of the yeast gcr1 mutant in terms of global genomic expression patterns. J Bacteriol 182:4970–4978
Lundblad V (1997) Currents protocols in molecular biology. Wiley, New York
Müller S, Boles E, May M, Zimmermann FK (1995) Different internal metabolites trigger the induction of glycolytic gene expression in Saccharomyces cerevisiae. J Bacteriol 177:4517–4519
Nishi K, Park CS, Pepper AE, Eichinger G, Innis MA, Holland MJ (1995) The GCR1 requirement for glycolytic gene expression is suppressed by dominant mutations in the SGC1 gene which encodes a novel basic-helix-loop-helix protein. Mol Cell Biol 15:2646–2653
Özcan S, Johnston M (1999) Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev 63:554–569
Prior C, Mamessier P, Fukuhara H, Chen XJ, Wésolowski-Louvel M (1993) The hexokinase gene is required for the transcriptional regulation of the glucose transporter gene RAG1 in Kluyveromyces lactis. Mol Cell Biol 13:3882–3889
Santangelo GM, Tornow J (1990) Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites Mol Cell Biol 10:859–862
Sato T, Lopez MC, Sugioka S, Jigami Y, Baker HV, Uemura H (1999) The E-box DNA binding protein Sgc1p suppresses the gcr2 mutation, which is involved in transcriptional activation of glycolytic genes in Saccharomyces cerevisiae. FEBS Lett 463:307–311
Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27
Thierry A, Fairhead C, Dujon B (1990) The complete sequence of the 8.2 kb segment left of MAT on chromosome III reveals five ORFs, including a gene for yeast ribokinase. Yeast 6:521–534
Tornow J, Zeng X, Gao W, Santangelo GM (1993) GCR1, a transcriptional activator in Saccharomyces cerevisiae complexes with RAP1 and can function without its DNA binding domain. EMBO J 12:2431–2437
Türkel S, Bisson LF (1999) Transcription of the HXT4 gene is regulated by Gcr1p and Gcr2p in the yeast S. cerevisiae. Yeast 15:1045–1057
Uemura H, Fraenkel DG (1990) gcr2, a new mutation affecting glycolytic gene expression in Saccharomyces cerevisiae. Mol Cell Biol 10:6389–6396
Uemura H, Jigami Y (1992) Role of GCR2 in transcriptional activation of yeast glycolytic genes. Mol Cell Biol 12:3834–3842
Wach A (1996) PCR-synthesis of marker cassettes with long flanking homology regions for gene disruption in S. cerevisiae. Yeast 12:259–265
Wésolowski-Louvel M, Goffrini P, Ferrero I, Fukuhara H (1992a) Glucose transport in the yeast Kluyveromyces lactis. I. Properties of an inducible low-affinity glucose transporter gene. Mol Gen Genet 33:89–96
Wésolowski-Louvel M, Prior C, Bornecque D, Fukuhara H (1992b) Rag- mutations involved in glucose metabolism in yeast: isolation and genetic characterization. Yeast 8:711–719
Zeng N, Deminoff SJ, Santangelo GM (1997) Specialized Rap1p/Gcr1p transcriptional activation through Gcr1p DNA contacts requires Gcr2p, as does hyperphosphorylation of Gcr1p. Genetics 147:495–506
Acknowledgements
H.N. was the recipient of a fellowship from the Ministère de la Recherche of France. We are grateful to Hiroshi Uemura for providing plasmids pL1617-9 and pL574-4. We thank Hiroshi Fukuhara and Steve Neil for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Breunig
Rights and permissions
About this article
Cite this article
Neil, H., Lemaire, M. & Wésolowski-Louvel, M. Regulation of glycolysis in Kluyveromyces lactis: role of KlGCR1 and KlGCR2 in glucose uptake and catabolism. Curr Genet 45, 129–139 (2004). https://doi.org/10.1007/s00294-003-0473-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00294-003-0473-5