Abstract
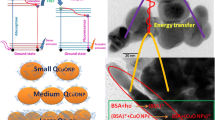
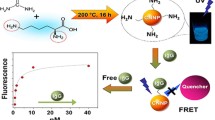
Among nanomaterials, we can now distinguish a special class called nanoflowers (NFs). These new nanostructures have aroused the interest of scientists due to the topographic features of nanolayers, the special location which allows a higher surface-to-volume ratio compared to classical spherical nanoparticles, thereby significantly increasing the efficiency of surface reactions for nanoflowers. The main value of nanoflowers is their action as enzyme stabilizers. A protein stability is usually enhanced by immobilization on a nanoflower surface through charge affinity and covalent bonds. The possibility of their use in vivo in biocatalysis, biosensors and medicine has been also investigated. We now report on the synthesis of two different nanoflowers: Cu nanoflowers and Fe3+ attached Cu nanoflowers and their interaction with two fluorescent probes, anilino-1-naphthalenesulfonic acid (ANS) and Fura 2, and two proteins, human serum albumin (HSA) and thrombin. Nanoflowers did not bind ANS, but bind efficiently to Fura 2 and both proteins. Modification of Cu–NFs by Fe3+ leads to significant changes in their binding capacity to fluorescent probe Fura 2 and both proteins. Their ability to bind fluorescent probe Fura 2 increased eightfold, and their ability to bind HSA and thrombin increased five times. Regarding Fe3+–Cu–NFs, a difference in binding between HSA and thrombin was found that can be explained by their structural features. Our data indicate the possibility of using studied nanoflowers for sorption of fluorescent probes and proteins.








Similar content being viewed by others
References
Cui J, Jia S (2017) Organic–inorganic hybrid nanoflowers: a novel host platform for immobilizing biomolecules. Coord Chem Rev. https://doi.org/10.1016/j.ccr.2017.09.008
Sassolas A, Blum LJ, Leca-Bouvier BD (2012) Immobilization strategies to develop enzymatic biosensors. Biotechnol Adv. https://doi.org/10.1016/j.biotechadv.2011.09.003
Datta S, Christena LR, Rajaram YRS (2013) Enzyme immobilization: an overview on techniques and support materials. 3 Biotech. https://doi.org/10.1007/s13205-012-0071-7
Kim J, Grate JW, Wang P (2008) Nanobiocatalysis and its potential applications. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2008.07.009
Ge J, Lu D, Liu Z, Liu Z (2009) Recent advances in nanostructured biocatalysts. Biochem Eng J. https://doi.org/10.1016/j.bej.2009.01.002
Luckarift HR, Spain JC, Naik RR, Stone MO (2004) Enzyme immobilization in a biomimetic silica support. Nat Biotechnol. https://doi.org/10.1038/nbt931
Mateo C, Grazu V, Palomo JM, Lopez-Gallego F, Fernandez-Lafuente R, Guisan JM (2007) Immobilization of enzymes on heterofunctional epoxy supports. Nat Protoc. https://doi.org/10.1038/nprot.2007.133
Ge J, Lei J, Zare RN (2012) Protein-inorganic hybrid nanoflowers. Nat Nanotechnol. https://doi.org/10.1038/nnano.2012.80
Tran TD, Il Kim M (2018) Organic-inorganic hybrid nanoflowers as potent materials for biosensing and biocatalytic applications. Biochip J. https://doi.org/10.1007/s13206-018-2409-7
Negahdary M, Heli H (2018) Applications of nanoflowers in biomedicine. Recent Pat Nanotechnol. https://doi.org/10.2174/1872210511666170911153428
Altinkaynak C, Tavlasoglu S, Özdemir N, Ocsoy I (2016) A new generation approach in enzyme immobilization: organic-inorganic hybrid nanoflowers with enhanced catalytic activity and stability. Enzyme Microb Technol. https://doi.org/10.1016/j.enzmictec.2016.06.011
Aydemir D, Geçili F, Özdemir N, UlusuNuray N (2020) A novel triple enzyme-inorganic hybrid nanoflower (TrpE@ihNF) with enhanced catalytic activity and stability. J Biosci Bioeng 129:679–686. https://doi.org/10.1016/j.jbiosc.2020.01.008
Zhu J, Wen M, Wen W, Du D, Zhang X, Wang S, Lin Y (2018) Recent progress in biosensors based on organic-inorganic hybrid nanoflowers. Biosens Bioelectron. https://doi.org/10.1016/j.bios.2018.08.058
Shcharbin D, Halets-Bui I, Abashkin V, Dzmitruk V, Loznikova S, Odabaşı M, Acet Ö, Önal B, Özdemir N, Shcharbina N, Bryszewska M (2019) Hybrid metal-organic nanoflowers and their application in biotechnology and medicine. Coll Surf B Biointerfaces 182:110354. https://doi.org/10.1016/j.colsurfb.2019.110354
Gurbuz F, Ceylan Ş, Odabaşi M, Codd GA (2016) Hepatotoxic microcystin removal using pumice embedded monolithic composite cryogel as an alternative water treatment method. Water Res. https://doi.org/10.1016/j.watres.2015.12.042
Önal B, Acet Ö, Dzmitruk V, Halets-Bui I, Shcharbin D, Özdemir N, Odabaşı M (2021) First protein affinity application of Cu2+-bound pure inorganic nanoflowers. Polym Bull. https://doi.org/10.1007/s00289-021-03557-5
Cui J, Zhao Y, Liu R, Zhong C, Jia S (2016) Surfactant-activated lipase hybrid nanoflowers with enhanced enzymatic performance. Sci Rep. https://doi.org/10.1038/srep27928
Bian H, Wang G, Cao M, Wang Z, Cui J (2020) Improved biodegradation of polyvinyl alcohol by hybrid nanoflowers of degrading enzymes from bacillus niacini. Korean J Chem Eng. https://doi.org/10.1007/s11814-020-0547-9
Li C, Zhao J, Zhang Z, Jiang Y, Bilal M, Jiang Y, Jia S, Cui J (2020) Self-assembly of activated lipase hybrid nanoflowers with superior activity and enhanced stability. Biochem Eng J. https://doi.org/10.1016/j.bej.2020.107582
Önal B, Acet Ö, Sanz R, Sanz-Pérez ES, Erdönmez D, Odabaşı M (2019) Co-evaluation of interaction parameters of genomic and plasmid DNA for a new chromatographic medium. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2019.09.068
Swain SK, Sarkar D (2013) Study of BSA protein adsorption/release on hydroxyapatite nanoparticles. Appl Surf Sci 286:99–103
Galdino FE, Picco AS, Sforca ML, Cardoso MB, Loh W (2020) Effect of particle functionalization and solution properties on the adsorption of bovine serum albumin and lysozyme onto silica nanoparticles. Coll Surf B Biointerfaces. https://doi.org/10.1016/j.colsurfb.2019.110677
Zhao L, Li L, Zhu C, Ghulam M, Qu F (2020) pH-responsive polymer assisted aptamer functionalized magnetic nanoparticles for specific recognition and adsorption of proteins. Anal Chim Acta. https://doi.org/10.1016/j.aca.2019.11.001
Serinbaş A, Önal B, Acet Ö, Özdemir N, Dzmitruk V, Halets-Bui I, Shcharbin D, Odabaşı M (2020) A new application of inorganic sorbent for biomolecules: IMAC practice of Fe3+-nano flowers for DNA separation. Mater Sci Eng C. https://doi.org/10.1016/j.msec.2020.111020
Luo YK, Song F, Wang XL, Wang YZ (2017) Pure copper phosphate nanostructures with controlled growth: a versatile support for enzyme immobilization. Cryst Eng Comm. https://doi.org/10.1039/c7ce00466d
Gulmez C, Altinkaynak C, Özdemir N, Atakisi O (2018) Proteinase K hybrid nanoflowers (P-hNFs) as a novel nanobiocatalytic detergent additive. Int J Biol Macromol. https://doi.org/10.1016/j.ijbiomac.2018.07.195
Schönbrunn E, Eschenburg S, Luger K, Kabsch W, Amrhein N (2000) Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.120120397
Matulis D, Baumann CG, Bloomfield VA, Lovrien RE (1999) 1-Anilino-8-naphthalene sulfonate as a protein conformational tightening agent. Biopolymers. https://doi.org/10.1002/(SICI)1097-0282(199905)49:6%3c451::AID-BIP3%3e3.0.CO;2-6
Grynkiewicz G, Poenie M, Tsien RY (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem 260(6):3440–3450
Marchi B, Burlando B, Panfoli I, Viarengo A (2000) Interference of heavy metal cations with fluorescent Ca2+ probes does not affect Ca2+ measurements in living cells. Cell Calcium. https://doi.org/10.1054/ceca.2000.0155
Curry S, Mandelkow H, Brick P, Franks N (1998) Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites. Nat Struct Biol. https://doi.org/10.1038/1869
Petitpas I, Petersen CE, Ha CE, Bhattacharya AA, Zunszain PA, Ghuman J, Bhagavan NV, Curry S (2003) Structural basis of albumin-thyroxine interactions and familial dysalbuminemic hyperthyroxinemia. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.1137188100
Zhang E, Tulinsky A (1997) The molecular environment of the Na+ binding site of thrombin. Biophys Chem. https://doi.org/10.1016/S0301-4622(96)02227-2
Di Cera E (2008) Thrombin. Mol Asp Med. https://doi.org/10.1016/j.mam.2008.01.001
Meloun B, Morávek L, Kostka V (1975) Complete amino acid sequence of human serum albumin. FEBS Lett. https://doi.org/10.1016/0014-5793(75)80242-0
Porath J, Carlsson J, Olsson I, Belfrage G (1975) Metal chelate affinity chromatography, a new approach to protein fractionation. Nature. https://doi.org/10.1038/258598a0
Hansen P, Andersson L, Lindeberg G (1996) Purification of cysteine-containing synthetic peptides via selective binding of the α-amino group to immobilised Cu2+ and Ni2+ ions. J Chromatogr A. https://doi.org/10.1016/0021-9673(95)00806-3
Arnold FH (1991) Metal-affinity separations: a new dimension in protein processing. Nat Biotechnol. https://doi.org/10.1038/nbt0291-151
Porath J, Olin B (1983) Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry. https://doi.org/10.1021/bi00276a015
Gutiérrez R, Martín Del Valle EM, Galán MA (2007) Immobilized metal-ion affinity chromatography: status and trends. Sep Purif Rev. https://doi.org/10.1080/15422110601166007
Shcharbin D, Klajnert B, Bryszewska M (2005) The effect of PAMAM dendrimers on human and bovine serum albumin at different pH and NaCl concentrations. J Biomater Sci Polym Ed 16:1081–1093. https://doi.org/10.1163/1568562054798518
Peters T (1996) All about albumin: biochemistry. Genetic and Medical Applications Elsevier, pp 133–187
Shakhbazau A, Shcharbin D, Seviaryn I, Goncharova N, Kosmacheva S, Potapnev M, Gabara B, Ionov M, Bryszewska M (2010) Use of polyamidoamine dendrimers to engineer BDNF-producing human mesenchymal stem cells. Mol Biol Rep 37:2003–2008. https://doi.org/10.1007/s11033-009-9651-y
Dzmitruk V, Apartsin E, Ihnatsyeu-Kachan A, Abashkin V, Shcharbin D, Bryszewska M (2018) Dendrimers show promise for siRNA and microrna therapeutics. Pharmaceutics. https://doi.org/10.3390/pharmaceutics10030126
Shcharbin D, Shcharbina N, Dzmitruk V, Pedziwiatr-Werbicka E, Ionov M, Mignani S, de la Mata FJ, Gómez R, Muñoz-Fernández MA, Majoral J-P, Bryszewska M (2017) Dendrimer-protein interactions versus dendrimer-based nanomedicine. Coll Surf B Biointerfaces. https://doi.org/10.1016/j.colsurfb.2017.01.041
Acknowledgements
This work was supported by the Technological Research Council of Turkey (TUBITAK) (Grant Number:118Z037) and by the State Committee of Science and Technology of Belarus and Belarussian Republican Foundation for Fundamental Research, grants B18TUB-001, B21TUB-001, B21KORG-001; by the Polish National Agency for Academic Exchange, grant EUROPARTNER, No. PPI/APM/2018/1/00007/U/001.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Halets-Bui, I., Dzmitruk, V., Abashkin, V. et al. Differences between Cu- and Fe–Cu nanoflowers in their interactions with fluorescent probes ANS and Fura-2 and proteins albumin and thrombin. Polym. Bull. 79, 5247–5259 (2022). https://doi.org/10.1007/s00289-021-03773-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00289-021-03773-z