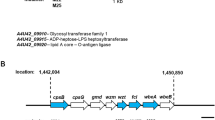
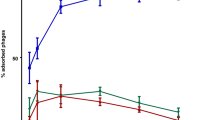
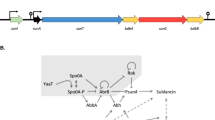
Abstract
Filamentous bacteriophage cf infects Xanthomonas axonopodis pv. citri, a serious plant pathogen which causes citrus canker. To understand the immunity regulation of bacteria infected with bacteriophage cf, we applied DNA shuffling to mutate the cf intergenic region. One of the immunity mutants, cf-m3 (NCBI Taxonomy ID: 3050368) expressed a 106–109 fold greater superinfection ability compared with wild type cf. Nine mutations were identified on the cf-m3 phage, four of which were located within the coding region of an open reading frame (ORF165) for a hypothetical repressor, PT, and five located upstream of the PT coding region. A set of phages with mutations to the predicted PT protein or the upstream coding region were generated. All showed similarly low superinfection efficiency to wild type cf and no superinfection ability on cf lysogens. The results indicate that rather than superinfection inhibition, the PT protein and the un-transcribed cis element function individually as positive regulators of cf superinfection immunity. Greater superinfection ability depends on the simultaneous presence of both elements. This work yields further insight into the possible control of citrus canker disease through phages that overcome host superinfection immunity.






Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article.
Code Availability
Not applicable.
References
Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK (2006) Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol 29(8):690–695. https://doi.org/10.1016/j.syapm.2006.08.001
Zhang Y, Jalan N, Zhou X, Goss E, Jones JB, Setubal JC, Deng X, Wang N (2015) Positive selection is the main driving force for evolution of citrus canker-causing Xanthomonas. ISME J 9:2128–2138. https://doi.org/10.1038/ismej.2015.15
Hay ID, Lithgow T (2019) Filamentous phages: masters of a microbial sharing economy. EMBO Rep 20:e47427. https://doi.org/10.15252/embr.201847427
Kuo TT, Chao YS, Lin YH, Lin BY, Liu LF, Feng TY (1987) Integration of the DNA of filamentous bacteriophage Cflt into the chromosomal DNA of its host. J Virol 61(1):60–65. https://doi.org/10.1128/jvi.61.1.60-65.1987
Shieh GJ, Charng YC, Yang BC, Tu J, Bau HJ, Kuo TT (1991) Identification and nucleotide sequence analysis of an open reading frame involved in high-frequency conversion of turbid to clear plaque mutants of filamentous phage Cf1t. Virology 185(1):316–322. https://doi.org/10.1016/0042-6822(91)90779-b
Cheng CM, Wang HJ, Bau HJ, Kuo TT (1999) The primary immunity determinant in modulating the lysogenic immunity of the filamentous bacteriophage cf. J Mol Biol 287(5):867–876. https://doi.org/10.1006/jmbi.1999.2651
Biggs KRH, Bailes CL, Scott L, Wichman HA, Schwartz EJ (2021) Ecological approach to understanding superinfection inhibition in bacteriophage. Viruses 13(7):1389. https://doi.org/10.3390/v13071389
Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, Maxwell KL (2016) Prophages mediate defense against phage infection through diverse mechanisms. The ISME J 10:2854–2866. https://doi.org/10.1038/ismej.2016.79
van Houte S, Buckling A, Westra ER (2016) Evolutionary ecology of prokaryotic immune mechanisms. Microbiol Mol Biol Rev 80(3):745–763. https://doi.org/10.1128/MMBR.00011-16
Tung SY, Kuo TT (1999) Requirement for phosphoglucose isomerase of Xanthomonas campestris in pathogenesis of citrus canker. Appl Environ Microbiol 65(12):5564–5570. https://doi.org/10.1128/AEM.65.12.5564-5573.1999
Maniatis T, Fritsch EF, Sambrook J, Fritsch EF, Sambrook J (1982) Molecular cloning: a laboratory manual, 1st edn. Cold Spring Harbor Laboratory, New York
Stemmer WP (1994) DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA 91:10747–10751. https://doi.org/10.1073/pnas.91.22.10747
Coco WM, Levinson WE, Crist MJ, Hektor HJ, Darzins A, Pienkos PT, Squires CH, Monticello DJ (2001) DNA suffling method for generating highly recombined genes and evolved enzymes. Nat Biotechnol 19:354–359. https://doi.org/10.1038/86744
Langlet J, Gaboriaud F, Gantzer C (2007) Effects of pH on plaque forming unit counts and aggregation of MS2 bacteriophage. J Appl Microbiol 103(5):1632–1638. https://doi.org/10.1111/j.1365-2672.2007.03396.x
Boratyn GM, Schaffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL (2012) Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12. https://doi.org/10.1186/1745-6150-7-12
Grissa I, Vergnaud G, Pourcel C (2007) CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35(2):W52-57. https://doi.org/10.1093/nar/gkm360
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415. https://doi.org/10.1093/nar/gkg595
Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50(W1):W276–W279. https://doi.org/10.1093/nar/gkac240
Brázda V, Kolomazník J, Lýsek J, Hároníková L, Coufal J, Št’astný J (2016) Palindrome analyser—a new web-based server for predicting and evaluating inverted repeats in nucleotide sequences. Biochem Biophys Res Commun 478(4):1739–1745. https://doi.org/10.1016/j.bbrc.2016.09.015
Li SC, Goto NK, Williams KA, Deber CM (1996) Alpha-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc Natl Acad Sci USA 93(13):6676–6681. https://doi.org/10.1073/pnas.93.13.6676
Melnikov S, Mailliot J, Rigger L, Neuner S, Shin BS, Yusupova G, Dever TE, Micura R, Yusupov M (2016) Molecular insights into protein synthesis with proline residues. EMBO Rep 17(12):1776–1784. https://doi.org/10.15252/embr.201642943
Wang HJ, Cheng CM, Wang CN, Kuo TT (1999) Transcription of the genome of the filamentous bacteriophage cf from both plus and minus DNA strands. Virology 256(2):228–232. https://doi.org/10.1006/viro.1999.9623
Briani F, Ghisotti D, Dehò G (2000) Antisense RNA-dependent transcription termination sites that modulate lysogenic development of satellite phage P4. Mol Microbiol 36(5):1124–1134. https://doi.org/10.1046/j.1365-2958.2000.01927.x
Forti F, Dragoni I, Briani F, Dehò G, Ghisotti D (2002) Characterization of the small antisense CI RNA that regulates bacteriophage P4 immunity. J Mol Biol 315(4):541–549. https://doi.org/10.1006/jmbi.2001.5274
Lewis D, Le P, Zurla C, Finzi L, Adhya S (2011) Multilevel autoregulation of lambda repressor protein CI by DNA looping in vitro. Proc Natl Acad Sci USA 108(36):14807–14812. https://doi.org/10.1073/pnas.1111221108
Forti F, Polo S, Lane KB, Six EW, Sironi G, Dehò G, Ghisotti D (1999) Translation of two nested genes in bacteriophage P4 controls immunity-specific transcription termination. J Bacteriol 181(17):5225–5233. https://doi.org/10.1128/jb.181.17.5225-5233.1999
Thomason LC, Morrill K, Murray G, Court C, Shafer B, Schneider TD, Court DL (2019) Elements in the lambda immunity region regulate phage development: beyond the “Genetic Switch.” Mol Microbiol 112(6):1798–1813. https://doi.org/10.1111/mmi.14394
Oppenheim AB, Kobiler O, Stavans J, Court DL, Adhya S (2005) Switches in bacteriophage lambda development. Annu Rev Genet 39:409–429. https://doi.org/10.1146/annurev.genet.39.073003.113656
van Beljouw SPB, Sanders J, Rodríguez-Molina A, Brouns SJJ (2023) RNA-targeting CRISPR-Cas systems. Nat Rev Microbiol 21(1):21–34. https://doi.org/10.1038/s41579-022-00793-y
Semenova E, Nagornykh M, Pyatnitskiy M, Artamonova II, Severinov K (2009) Analysis of CRISPR system function in plant pathogen Xanthomonas oryzae. FEMS Microbiol Lett 296(1):110–116. https://doi.org/10.1111/j.1574-6968.2009.01626.x
van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ (2009) CRISPR-based adaptive and heritable immunity in prokaryotes. Trend Biochem Sci 34(8):401–407. https://doi.org/10.1016/j.tibs.2009.05.002
Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. https://doi.org/10.1038/nature09523
Davidson AR, Lu WT, Stanley SY, Wang J, Mejdani M, Trost CN, Hicks BT, Lee J, Sontheimer EJ (2020) Anti-CRISPRs: protein inhibitors of CRISPR-Cas systems. Annu Rev Biochem 89:309–332. https://doi.org/10.1146/annurev-biochem-011420-111224
Haft DH, Selengut J, Mongodin EF, Nelson KE (2005) A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol 1:e60. https://doi.org/10.1371/journal.pcbi.0010060
Vasu K, Rao DN, Nagaraja V (2019) Restriction-modification systems. In: Schmidt TM (ed) Encyclopedia of microbiology, 4th edn. Academic Press, Cambridge, pp 102–109
Nakayama Y, Kobayashi I (1998) Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc Natl Acad Sci USA 95(11):6442–6447. https://doi.org/10.1073/pnas.95.11.6442
Murphy J, Klumpp J, Mahony J, O’Connell-Motherway M, Nauta A, van Sinderen D (2014) Methyltransferases acquired by lactococcal 936-type phage provide protection against restriction endonuclease activity. BMC Genom 15(1):831. https://doi.org/10.1186/1471-2164-15-831
da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LM, do Amaral AM, Bertolini MC, Camargo LE, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Ciapina LP, Cicarelli RM, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJ, Ferreira RC, Ferro MI, Formighieri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Leite RP, Lemos EG, Lemos MV, Locali EC, Machado MA, Madeira AM, Martinez-Rossi NM, Martins EC, Meidanis J, Menck CF, Miyaki CY, Moon DH, Moreira LM, Novo MT, Okura VK, Oliveira MC, Oliveira VR, Pereira HA, Rossi A, Sena JA, Silva C, de Souza RF, Spinola LA, Takita MA, Tamura RE, Teixeira EC, Tezza RI, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP, (2002) Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417(6887):459–463. https://doi.org/10.1038/417459a
Szczesny R, Jordan M, Schramm C, Schulz S, Cogez V, Bonas U, Büttner D (2010) Functional characterization of the Xcs and Xps type II secretion systems from the plant pathogenic bacterium Xanthomonas campestris pv vesicatoria. New Phytol 187(4):983–1002. https://doi.org/10.1111/j.1469-8137.2010.03312.x
Furuta Y, Abe K, Kobayashi I (2010) Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res 38(7):2428–2443. https://doi.org/10.1093/nar/gkp1226
Choi Y, Kim N, Mannaa M, Kim H, Park J, Jung H, Han G, Lee HH, Seo YS (2020) Characterization of type VI secretion system in Xanthomonas oryzae pv. oryzae and its role in virulence to rice. Plant Pathol J 36(3):289–296. https://doi.org/10.5423/PPJ.NT.02.2020.0026
Weigele P, Raleigh EA (2016) Biosynthesis and function of modified bases in bacteria and their viruses. Chem Rev 116(20):12655–12687. https://doi.org/10.1021/acs.chemrev.6b00114
Huang LH, Farnet CM, Ehrlich KC, Ehrlich M (1982) Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res 10(5):1579–1591. https://doi.org/10.1093/nar/10.5.1579
Nakayinga R, Makumi A, Tumuhaise V, Tinzaara W (2021) Xanthomonas bacteriophages: a review of their biology and biocontrol applications in agriculture. BMC Microbiol 21(1):291. https://doi.org/10.1186/s12866-021-02351-7
Seong HJ, Park HJ, Hong E, Lee SC, Sul WJ, Han SW (2016) Methylome analysis of two Xanthomonas spp. using single-molecule real-time sequencing. Plant Pathol J 32(6):500–507. https://doi.org/10.5423/PPJ.FT.10.2016.0216
Xiao CL, Xie SQ, Xie QB, Liu ZY, Xing JF, Ji KK, Tao J, Dai LY, Luo F (2018) N6-Methyladenine DNA modification in Xanthomonas oryzae pv. oryzicola genome. Sci Rep 8(1):16272. https://doi.org/10.1038/s41598-018-34559-5
Park HJ, Jung B, Lee J, Han SW (2019) Functional characterization of a putative DNA methyltransferase, EadM, in Xanthomonas axonopodis pv. glycines by proteomic and phenotypic analyses. Sci Rep 9(1):2446. https://doi.org/10.1038/s41598-019-38650-3
Millman A, Melamed S, Leavitt A, Doron S, Bernheim A, Hör J, Garb J, Bechon N, Brandis A, Lopatina A, Ofir G, Hochhauser D, Stokar-Avihail A, Tal N, Sharir S, Voichek M, Erez Z, Ferrer JLM, Dar D, Kacen A, Amitai G, Sorek R (2022) An expanded arsenal of immune systems that protect bacteria from phages. Cell Host Microbe 30(11):1556-1569.e5. https://doi.org/10.1016/j.chom.2022.09.017
Huiting E, Bondy-Denomy J (2023) Defining the expanding mechanisms of phage-mediated activation of bacterial immunity. Curr Opin Microbiol 74:102325. https://doi.org/10.1016/j.mib.2023.102325
Kranzusch PJ (2023) Editorial overview: evolution of antiviral defense. Curr Opin Microbiol 2023(75):102352. https://doi.org/10.1016/j.mib.2023.102352
Doron S, Melamed S, Ofir G, Leavitt A, Lopatina A, Keren M, Amitai G, Sorek R (2018) Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359(6379):eaar4120. https://doi.org/10.1126/science.aar4120
Gao L, Altae-Tran H, Böhning F, Makarova KS, Segel M, Schmid-Burgk JL, Koob J, Wolf YI, Koonin EV, Zhang F (2020) Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 369(6507):1077–1084. https://doi.org/10.1126/science.aba0372
Bernheim A, Sorek R (2020) The pan-immune system of bacteria: antiviral defence as a community resource. Nat Rev Microbiol 18(2):113–119. https://doi.org/10.1038/s41579-019-0278-2
Tal N, Morehouse BR, Millman A, Stokar-Avihail A, Avraham C, Fedorenko T, Yirmiya E, Herbst E, Brandis A, Mehlman T, Oppenheimer-Shaanan Y, Keszei AFA, Shao S, Amitai G, Kranzusch PJ, Sorek R (2021) Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cell 184(23):5728-5739.e16. https://doi.org/10.1016/j.cell.2021.09.031
Acknowledgements
We would like to thank Dr. Pelle Stolt for critical reading and editing of the manuscript and Miss Shan-Hus Lin for helpful assistance. Without their care and consideration, this work would not have been possible.
Funding
This work was financially supported by Chi-Tzu University (TCIRP98005-03).
Author information
Authors and Affiliations
Contributions
CMC designed the study. CCY and ZYW performed the experiments. All authors were involved in the analysis. CMC wrote the main manuscript text.
Corresponding author
Ethics declarations
Conflict of interests
The authors have not disclosed any competing interests.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, CC., Wang, ZY. & Cheng, CM. Insights into Superinfection Immunity Regulation of Xanthomonas axonopodis Filamentous Bacteriophage cf. Curr Microbiol 81, 42 (2024). https://doi.org/10.1007/s00284-023-03539-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03539-y