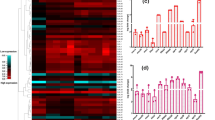
Abstract
Staphylococcus aureus is a global pathogen and is responsible for causing severe life-threatening infections. The current study was designed to investigate transcriptional expression of different core, regulatory, and accessory genes within vanB operon under differential exposure of vancomycin and teicoplanin. Four isolates selected for the study, were confirmed to harbour vanB gene in which three isolates showed MIC breakpoint above 16 µg/ml and one isolate above 8 µg/ml against vancomycin while teicoplanin showed higher MIC breakpoint as compared to vancomycin. Antibiotic susceptibility results showed that these isolates were susceptible towards imipenem and linezolid. Transcriptional expressional analysis of the core gene of vanB operon showed that expression of vanB is increased under vancomycin stress but is inversely proportional to increase in the concentration of the vancomycin while under teicoplanin stress the expression of vanB showed no significant pattern. Similar expressional pattern was found for vanH gene for both the glycopeptides. In case of vanX, expression was significantly increased at 1 µg/ml exposure of vancomycin, however, no pattern could be observed in case of teicoplanin stress. In case of regulatory gene, vanR, significant increase in expression was observed under vancomycin and teicoplanin stress of 1 µg/ml, however vanS, showed significant increase in the expression under 1 µg/ml of vancomycin. The accessory gene, vanY showed marginal increase in expression under both the antibiotic, while in case of vanW, the expressional pattern was found to be inversely proportional to the increasing antibiotic concentration.




Similar content being viewed by others
Data Availability
All the data will be made available on request.
Code Availability
Not applicable.
References
Jahanshahi A, Zeighami H, Haghi F (2018) Molecular characterization of methicillin and vancomycin resistant Staphylococcus aureus strains isolated from hospitalized patients. Microb Drug Resist 24:1529–1536. https://doi.org/10.1089/mdr.2018.0069
George SK, Suseela MR, El Safi S, Elnagi EA, Al-Naam YA, Adam AAM, Ks HK (2021) Molecular determination of van genes among clinical isolates of enterococci at a hospital setting. Saudi J Biol Sci 28:2895–2899. https://doi.org/10.1016/j.sjbs.2021.02.022
Saha B, Singh AK, Ghosh A, Bal M (2008) Identification and characterization of a vancomycin-resistant Staphylococcus aureus isolated from Kolkata (South Asia). J Med Microbiol 57:72–79. https://doi.org/10.1099/jmm.0.47144-0
Pootoolal J, Neu J, Wright GD (2002) Glycopeptide antibiotic resistance. Annu Rev Pharmacol Toxicol 42:381. https://doi.org/10.1146/annurev.pharmtox.42.091601.142813
Guffey AA, Loll PJ (2021) Regulation of resistance in vancomycin-resistant enterococci the VanRS two-component system. Microorganisms 9:2026. https://doi.org/10.3390/microorganisms9102026
CLSI 2020. Performance Standards for Antimicrobial Susceptibility Testing 30th ed. CLSI supplement M100.Wayne, PA: Clinical and Laboratory Standards Institute
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Ziasistani M, Shakibaie MR, Kalantar-Neyestanaki D (2019) Genetic characterization of two vancomycin-resistant Staphylococcus aureus isolates in Kerman. Iran Infect Drug Resist 12:1869. https://doi.org/10.2147/IDR.S205596
Saadat S, Solhjoo K, Norooz-Nejad MJ, Kazemi A (2014) VanA and vanB positive vancomycin-resistant Staphylococcus aureus among clinical isolates in Shiraz. South Iran Oman Med J 29:335. https://doi.org/10.5001/omj.2014.90
Hill CM, Krause KM, Lewis SR, Blais J, Benton BM, Mammen M, Janc JW (2010) Specificity of induction of the vanA and vanB operons in vancomycin-resistant enterococci by telavancin. Antimicrob Agents Chemother 54:2814–2818. https://doi.org/10.1128/AAC.01737-09
Sadowy E (2021) Mobile genetic elements beyond the VanB-resistance dissemination among hospital-associated enterococci and other Gram-positive bacteria. Plasmid 114:102558. https://doi.org/10.1016/j.plasmid.2021.102558
Evers S, Courvalin P (1996) Regulation of VanB-type vancomycin resistance gene expression by the VanS (B)-VanR (B) two-component regulatory system in Enterococcus faecalis V583. J Bacteriol 178:1302–1309. https://doi.org/10.1128/jb.178.5.1302-1309.1996
Bamigboye BT, Olowe OA, Taiwo SS (2018) Phenotypic and molecular identification of vancomycin resistance in clinical Staphylococcus aureus isolates in Osogbo, Nigeria. Eur J Microbiol Immunol 8:25–30. https://doi.org/10.1556/1886.2018.00003
Cong Y, Yang S, Rao X (2020) Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J Adv Res 21:169–176. https://doi.org/10.1016/j.jare.2019.10.005
Ribeiro T, Santos S, Marques MIM, Gilmore M, Lopes MDFS (2011) Identification of a new gene, vanV, in vanB operons of Enterococcus faecalis. Int J Antimicrob Agents 37:554–557. https://doi.org/10.1016/j.ijantimicag.2011.01.024
Bisicchia P, Bui NK, Aldridge C, Vollmer W, Devine KM (2011) Acquisition of VanB-type vancomycin resistance by Bacillus subtilis: the impact on gene expression, cell wall composition and morphology. Mol Microbiol 81:157–178. https://doi.org/10.1111/j.1365-2958.2011.07684.x
Foucault ML, Depardieu F, Courvalin P, Grillot-Courvalin C (2010) Inducible expression eliminates the fitness cost of vancomycin resistance in enterococci. Proc Natl Acad Sci 107:16964–16969. https://doi.org/10.1073/pnas.1006855107
Acknowledgements
The authors would like to thank Council of Scientific and Industrial Research, Government of India [letter no. 27(0321)/17/EMR-II dated 12-04-2017] for providing financial support.
Funding
The study was financially supported by CSIR project, Government of India [letter no. 27(0321)/17/EMR-II dated 12–04-2017].
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by MH, JW, KN, DB, KMS, DDC, AB. The first draft of the manuscript was written by MH and AB. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have not disclosed any competing interests.
Ethical Approval
The study was ethically approved by Institutional ethical committee Assam University Silchar vide resolution 3(Sl.no 5) in the meeting on 09.04.2018.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hazarika, M., Wangkheimayum, J., Nath, K. et al. Transcriptional Response of vanB Operon in Staphylococcus aureus Against Vancomycin and Teicoplanin Stress. Curr Microbiol 80, 275 (2023). https://doi.org/10.1007/s00284-023-03389-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03389-8