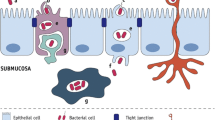
Abstract
Salmonella enterica is one of the foodborne pathogens that can infect humans, spreading from one person to another by contaminated food and water. To identify the pathogenic S. enterica from the contaminated food product, culture-based and molecular identifications, drug resistance profiling, virulence and genetic traits of the strains have been used. Herein, different animal products was subjected to screen for S. enterica prevalence, pathogenic characterization and compared with clinical Salmonella isolates (human). A total of 173 isolates from animal products and 51 isolates from clinical samples were collected. S. Typhi, S. Agona and S. Ohio were predominant serovars in blood, stool and different animal products. Both, clinical [37% (n = 19/51)] and animal product-associated isolates [21% (n = 37/173)] expressed their highest resistance to nalidixic acid. Thirty-one percentage of (n = 16/51) clinical isolates and 12% (n = 21/173) animal food-associated isolates were resistant to multiple classes of antibiotics. Class 1 integrons encoded by S. Typhi, S. Infantis and S. Emek were screened for sequence analysis, the result revealed that the cassettes encoded-aminoglycoside acetyltransferase and dihydrofolate reductase enzymes. Salmonella pathogenicity island-1 encoded-hilA gene was detected most frequently in all the isolates. PFGE profile revealed the genetic traits of the isolates which were closely linked with antibiotic-resistant properties and virulent characteristics. Only S. Enteritidis, collected from different samples had clonal similarities. In summary, drug-resistant pathogenic Salmonella prevalence was observed in the animal product that could be an important alarm to consumers with the risk of enteric fever and it causes the potential risk to public health.




Similar content being viewed by others
Data Availability
The integron gene sequencing data of the Salmonella enterica was submitted to NCBI data bse and the accession number is KP998196, KP742968 and KP728380.
Code Availability
Not applicable.
References
Wang B, Park B (2020) Immunoassay biosensing of foodborne pathogens with surface plasmon resonance imaging: a review. J Agric Food Chem 68(46):12927–12939. https://doi.org/10.1021/acs.jafc.0c02295
McAnearney S, McCall D (2015) Salmonella Osteomyelitis Ulst. Med J 84(3):171–172
Kern WV, Rieg S (2020) Burden of bacterial bloodstream infection—a brief update on epidemiology and significance of multidrug-resistant pathogens. Clin Microbiol Infect 26:151–157. https://doi.org/10.1016/j.cmi.2019.10.031
Van den Bogaart L, Lang BM, Neofytos D, Egli A, Walti LN, Boggian K, Garzoni C et al (2022) Epidemiology and outcomes of medically attended and microbiologically confirmed bacterial foodborne infections in solid organ transplant recipients. Am J Transplant 22(1):199–209. https://doi.org/10.1111/ajt.16831
Indrajith S, Athmanathan B, Subbaraj D K, Meganathan V, Arockiaraj JEE, Woodrowwilson JE, Karuppannan S (2015) Incidence and antibiotic resistant profiles of pathogenic Salmonella spp. from different environmental and food samples. Int J Microbiol Immunol Res 3(6): 76–83.
Besser JM (2018) Salmonella epidemiology: a whirlwind of change. Food Microbiol 71:55–59. https://doi.org/10.1016/j.fm.2017.08.018
Beyene AM, Gezachew M, Mengesha D, Yousef A, Gelaw B (2022) Prevalence and drug resistance patterns of Gram-negative enteric bacterial pathogens from diarrheic patients in Ethiopia: a systematic review and meta-analysis. PLoS ONE 17(3):e0265271. https://doi.org/10.1371/journal.pone.0265271
De Cesare A (2018) Salmonella in foods: a re-emerging problem. Adv Food Nutr Res 86:137–179. https://doi.org/10.1016/bs.afnr.2018.02.007
Popa GL, Popa MI (2020) Salmonella spp. Infection—a continuous threat worldwide. GERMS 11(1):88–96. https://doi.org/10.18683/germs.2021.1244
Hensel M (2004) Evolution of pathogenicity islands of Salmonella enterica. Int J Med Microbiol 294:95–102. https://doi.org/10.1016/j.ijmm.2004.06.025
Agbor TA, McCormick BA (2011) Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol 13:1858–1869. https://doi.org/10.1111/j.1462-5822.2011.01701.x
Kim JS, Liu L, Davenport B, Kant S, Morrison TE, Vazquez-Torres A (2022) Oxidative stress activates transcription of Salmonella pathogenicity island-2 genes in macrophages. J Biol Chem. https://doi.org/10.1016/j.jbc.2022.102130
Fluit AC (2005) Towards more virulent and antibiotic-resistant Salmonella? FEMS Microbiol Immunol 43:1–11. https://doi.org/10.1016/j.femsim.2004.10.007
Chantharothaiphaichit T, Phongaran D, Angkittitrakul S, Aunpromma S, Chuanchuen R (2022) Clinically healthy household dogs and cats as carriers of multidrug-resistant Salmonella enterica with variable R plasmids. J Med Microbiol 71(2):001488. https://doi.org/10.1099/jmm.0.001488
World Health Organization (2017) Global framework for development & stewardship to combat antimicrobial resistance—draft roadmap. World Health Organization, Geneva
Parry CM, Threlfall EJ (2008) Antimicrobial resistance in typhoidal and nontyphoidal Salmonellae. Curr Opin Infect Dis 21:531–538. https://doi.org/10.1097/QCO.0b013e32830f453a
Sharvani R, Hemavathi DK, Shenoy P, Sarmah P (2016) Antibiogram of Salmonella isolates: time to consider antibiotic salvage. J Clin Diagn Res 10: DC06–DC08. https://doi.org/10.7860/JCDR/2016/18102.7753.
Teng L, Liao S, Zhou X, Jia C, Feng M, Pan H, Ma Z, Yue M (2022) Prevalence and genomic investigation of multidrug-resistant Salmonella isolates from companion animals in Hangzhou, China. Antibiotics 11(5):625. https://doi.org/10.3390/antibiotics11050625
Mikoleit ML (2010) WHO Global Foodborne Infections Network. A WHO network building capacity to detect, control and prevent foodborne and other enteric infections from farm to table‖ Laboratory Protocol: isolation of Salmonella and Shigella from Faecal specimens”. Enteric diseases laboratory branch, Centers for Disease Control and Prevention. Atlanta, GA. 2010 Nov 19.
Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galan JE, Ginocchio C, Curtiss Iii R, Gyles CL (1992) Amplification of an InvA gene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes 6(4):271–279. https://doi.org/10.1016/0890-8508(92)90002-F
Grimont PA, Weill FX (2007) Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella 9:1–66
CLSI (2017) Performance standards for antimicrobial susceptibility testing. CLSI supplement M100.
Pazhani GP, Chakraborty S, Fujihara K, Yamasaki S, Ghosh A, Nair GB, Ramamurthy T (2011) QRDR mutations, efflux system & antimicrobial resistance genes in enterotoxigenic Escherichia coli isolated from an outbreak of diarrhoea in Ahmedabad. India Indian J Med Res 134:214–223
Bhowmick PP, Devegowda D, Karunasagar I (2011) Virulotyping of seafood associated Salmonella enterica subsp. enterica isolated from Southwest coast of India. Biotechnol Bioinforma Bioeng 1:63–69
Centers for Disease Control and Prevention (2013) Standard operating procedure for PulseNet PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. Centers for Disease Control and Prevention, Atlanta.
Ferrari RG, Rosario DKA, Cunha-Neto A, Mano SB, Figueiredo EES, Conte-Junior CA (2019) Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl Environ Microbiol 85:14. https://doi.org/10.1128/AEM.00591-19
Ramtahal MA, Amoako DG, Akebe ALK, Somboro AM, Bester LA, Essack SY (2022) A public health insight into Salmonella in poultry in Africa: a review of the past decade: 2010–2020. Microb Drug Resist 28(6):710–733. https://doi.org/10.1089/mdr.2021.0384
Abhijit A, Sunita N (2011) The study of salmonellosis with reference to Salmonella Typhi in enteric fever patients. J Clin Diagn Res 5:467–469
Wattal C, Goel N (2020) Pediatric blood cultures and antibiotic resistance: an overview. Indian J Pediatr 87:125–131. https://doi.org/10.1007/s12098-019-03123-y
Kumar Y, Gupta N, Vaish VB, Gupta S (2016) Distribution trends & antibiogram pattern of Salmonella enterica serovar Newport in India. Indian J Med Res 144:82–86. https://doi.org/10.4103/0971-5916.193293
Helmuth IG, Espenhain L, Ethelberg S, Jensen T, Kjeldgaard J, Litrup E, Schjorring S, Muller L (2019) An outbreak of monophasic Salmonella typhimurium associated with raw pork sausage and other pork products, Denmark 2018–19. Epidemiol. Infect. https://doi.org/10.1017/S0950268819002073.
Hoffmann M, Miller J, Melka D, Allard MW, Brown EW, Pettengill JB (2020) Temporal dynamics of Salmonella enterica subsp. enterica Serovar Agona isolates from a recurrent multistate outbreak. Front. Microbiol 11:478. https://doi.org/10.3389/fmicb.2020.00478.
Vázquez X, Fernández J, Hernáez S, Rodicio R, Rodicio MR (2022) Plasmid-Mediated Quinolone Resistance (PMQR) in Two Clinical Strains of Salmonella enterica Serovar Corvallis. Microorganisms 10(3):579. https://doi.org/10.3390/microorganisms10030579
Saravanan S, Purushothaman V, Murthy TR, Sukumar K, Srinivasan P, Gowthaman V, Balusamy M et al (2015) Molecular epidemiology of nontyphoidal Salmonella in poultry and poultry products in India: implications for human health. Indian J Microbiol 55:319–326. https://doi.org/10.1007/s12088-015-0530-z
Kalambhe DG, Zade NN, Chaudhari SP, Shinde SV, Khan W, Patil AR (2016) Isolation, antibiogram and pathogenicity of Salmonella spp. recovered from slaughtered food animals in Nagpur region of Central India. Vet World 9:176–181. https://doi.org/10.14202/vetworld.2016.176-181
Adesiji YO, Shivakumaraswamy SK, Deekshit VK, Kallappa GS, Karunasagar I (2017) Molecular characterization of antimicrobial multi-drug resistance in non-typhoidal Salmonellae from chicken and clam in Mangalore. India Indian J Med Res 32:237–244
Karabasanavar N, Sivaraman GK, SPS., Nair AS, Vijayan A, Rajan V, PS G (2022) Non-diarrhoeic pigs as source of highly virulent and multidrug-resistant non-typhoidal Salmonella. Braz J Microbiol. https://doi.org/10.1007/s42770-022-00700-w.
Taneja N, Appannanavar SB, Kumar A, Varma G, Kumar Y, Mohan B, Sharma M (2014) Serotype profile and molecular characterization of antimicrobial resistance in non-typhoidal Salmonella isolated from gastroenteritis cases over nine years. J Med Microbiol 63(1):66–73. https://doi.org/10.1099/jmm.0.061416-0
Kumar Y, Singh V, Kumar G, Gupta NK, Tahlan AK (2019) Serovar diversity of Salmonella among poultry. Indian J Med Res 150:92–95. https://doi.org/10.4103/ijmr.IJMR_1798_17
Killalea D, Ward LR, Roberts D, De Louvois J, Sufi F, Stuart JM, Wall PG et al (1996) International epidemiological and microbiological study of outbreak of Salmonella Agona infection from a ready to eat savoury snack–I: England and Wales and the United States. BMJ 313(7065):1105–1107. https://doi.org/10.1136/bmj.313.7065.1105
Sharma NC, Kumar D, Sarkar A, Chowdhury G, Mukhopadhyay AK, Ramamurthy T (2020) Prevalence of multidrug resistant Salmonellae with increasing frequency of Salmonella enterica serovars Kentucky and Virchow among hospitalized diarrheal cases in and around Delhi. India Jpn J Infect Dis 73:119–123. https://doi.org/10.7883/yoken.JJID.2019.063
Kumar R, Surendran PK, Thampuran N (2009) Analysis of antimicrobial resistance and plasmid profiles in Salmonella serovars associated with tropical seafood of India. Foodborne Pathog Dis 6(5):621–625. https://doi.org/10.1089/fpd.2008.0252
Nieto PA, Pardo-Roa C, Salazar-Echegarai FJ, Tobar HE, Coronado-Arrázola I, Riedel CA, Kalergis AM, Bueno SM (2016) New insights about excisable pathogenicity islands in Salmonella and their contribution to virulence. Microbes Infect 18:302–309. https://doi.org/10.1016/j.micinf.2016.02.001
Parvathi A, Vijayan J, Murali G, Chandran P (2011) Comparative virulence genotyping and antimicrobial susceptibility profiling of environmental and clinical Salmonella enterica from Cochin. India Curr Microbiol 62:21–26. https://doi.org/10.1007/s00284-010-9665-7
Dinjus U, Hanel I, Muller W, Bauerfeind R, Helmuth R (1997) Detection of the induction of Salmonella enterotoxin gene expression by contact with epithelial cells with RT-PCR. FEMS Microbiol Lett 146:175–179. https://doi.org/10.1111/j.1574-6968.1997.tb10189.x
Nair VTD, Venkitanarayanan K, Kollanoor Johny A (2018) Antibiotic-resistant Salmonella in the food supply and the potential role of antibiotic alternatives for control. Foods 7(10):167. https://doi.org/10.3390/foods7100167
Angelakis E (2017) Weight gain by gut microbiota manipulation in productive animals. Microb Pathog 106:162–170. https://doi.org/10.1016/j.micpath.2016.11.002
Rodríguez I, Martín MC, Mendoza MC and Rodicio MR (2006) Class 1 and class 2 integrons in non-prevalent serovars of Salmonella enterica: structure and association with transposons and plasmids. J Antimicrob Chemother 58(6):1124–1132. https://doi.org/10.1093/jac/dkl400.
Indrajith S, Mukhopadhyay AK, Chowdhury G et al (2021) Molecular insights of Carbapenem resistance Klebsiella pneumoniae isolates with focus on multidrug resistance from clinical samples. J Infect Public Health 14(1):131–138. https://doi.org/10.1016/j.jiph.2020.09.018
Thong KL, Ngeow YF, Altwegg M, Navaratnam P, Pang T (1995) Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol 33:1070–1074. https://doi.org/10.1128/jcm.33.5.1070-1074.1995
Acknowledgements
The authors wish to thank K.A.P. Viswanathan Medical College and Hospital, Tiruchirappalli, Tamil Nadu for providing the clinical isolates. We wish to thank Department of Bacteriology, National Institute of Cholera and Enteric Diseases (NICED) Beliaghata, Kolkata, India for providing lab facilities and support to do the serotyping and PFGE analysis. Also, The author wish to thank Department of Molecular Microbiology, School of Biotechnology, Madurai Kamaraj University for providing the lab facilities for the entire work.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
SI was responsible for sample collection, isolation, identification and characterization of the Salmonella isolates as well as manuscript preparation, SI and SN2 performed the Serotyping and PFGE analysis of the strains. SI, SN1 ad ST conceptualized and developed the project. SI, SN1 and ST contributed in the drafting, editing/proof reading and finalizing the manuscript. All authors read and approved the fnal manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conficts of interest.
Ethics Approval
This paper describes the prevalence and clonal diversity analysis of the bacterial isolate from human samples. The Ethics Committees of Institutional Bio-Safcty Committce (IBSC), Mahendra College of Arts & Science, Namakal has been reviewed and approved the proposed study by the IBSC on the meeting held on 20th March 2018 (IBSC/2018/03).
Consent to Participate
SI was responsible for sample collection, isolation, identification and characterization of the Salmonella isolates as well as manuscript preparation, SI and SN2 performed the Serotyping and PFGE analysis of the strains. SI, SN1 ad ST conceptualized and developed the project. SI, SN1 and ST contributed in the drafting, editing/proof reading and finalizing the manuscript. All authors read and approved the fnal manuscript.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Indrajith, S., Natarajan, S., Thangasamy, S. et al. Drug Resistance, Characterization and Phylogenetic Discrepancy of Salmonella enterica Isolates from Distinct Sources. Curr Microbiol 80, 314 (2023). https://doi.org/10.1007/s00284-023-03343-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-023-03343-8