Abstract
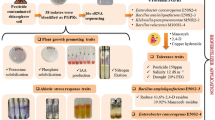
Disha A (Bacillus cereus) and Disha B (Bacillus safensis) were isolated from pesticide-infested agricultural field and showed tolerance against pesticides, heavy metals, and antibiotics. The isolates exhibited PGPR activities in vitro as well as in field conditions in lentil (Lens culinaris) and cow pea (Vigna unguiculata). Both the Bacillus species could not be grown in mineral salt medium but efficiently grown in the media supplemented with pesticide (imidacloprid/carbendazim) demonstrating the utilization of pesticide as carbon/nitrogen source. The HPLC studies exhibited the pesticide (imidacloprid/carbendazim) degradation by both the bacteria. B. safensis showed better degradation of carbendazim (88.93%) and imidacloprid (82.48%) than that of B. cereus 78.07% and 49.12%, respectively. The bacterial isolates showed high concentration of heavy metal tolerance viz. lead, molybdenum, cadmium, copper, cobalt, and zinc, except mercury. Both the bacteria possessed single plasmid. The plasmid-cured isolates of B. cereus did not tolerate any pesticide, whereas that of B. safensis tolerated all the pesticides, like wild strains. The plasmid curing experiments did not affect the heavy metal tolerance ability of both the bacteria indicating the genomic nature of heavy metal tolerance genes, whereas pesticide resistance genes are plasmid-dependent in B. cereus but genomic in B. safensis.





Similar content being viewed by others
Availability of Data and Material
Available in supplementary materials. The other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code Availability
Not applicable.
Abbreviations
- HPLC:
-
High-performance liquid chromatography
- LB:
-
Luria Bertani
- MS:
-
Mineral salt
- MSM:
-
Mineral salt medium
- MIC:
-
Minimum inhibitory concentration
- NB:
-
Nutrient broth
- NA:
-
Nutrient agar
- PGPR:
-
Plant growth-promoting rhizobacteria
- PGPF:
-
Plant growth-promoting fungi
References
Zhao Q, Yue S, Bilal M, Hu H, Wang W, Zhang X (2017) Comparative genomic analysis of 26 Sphingomonas and Sphingobium strains: dissemination of bioremediation capabilities, biodegradation potential and horizontal gene transfer. Sci Total Environ 609:1238–1247. https://doi.org/10.1016/j.scitotenv.2017.07.249
Jacob JM, Karthik C, Saratale RG, Kumar SS, Prabakar D, Kadirvelu K et al (2018) Biological approaches to tackle heavy metal pollution: a survey of literature. J Environ Manag 217:56–70. https://doi.org/10.1016/j.jenvman.2018.03.077
Barrios-Estrada C, de Jesús Rostro-Alanis M, Muñoz-Gutiérrez BD, Iqbal HM, Kannan S, Parra-Saldivar R (2018) Emergent contaminants: endocrine disruptors and their laccase-assisted degradation—a review. Sci Total Environ 612:1516–1531. https://doi.org/10.1016/j.scitotenv.2017.09.013
Rasheed T, Bilal M, Nabeel F, Iqbal HM, Li C, Zhou Y (2018) Fluorescent sensor based models for the detection of environmentally-related toxic heavy metals. Sci Total Environ 615:476–485. https://doi.org/10.1016/j.scitotenv.2017.09.126
Bilal M, Adeel M, Rasheed T, Zhao Y, Iqbal HM (2019) Emerging contaminants of high concern and their enzyme-assisted biodegradation—a review. Environ Int 124:336–353. https://doi.org/10.1016/j.envint.2019.01.011
Liu L, Bilal M, Duan X, Hafiz MN, Iqbal HMN (2019) Mitigation of environmental pollution by genetically engineered bacteria—current challenges and future perspectives. Sci Total Environ 667:444–454. https://doi.org/10.1016/j.scitotenv.2019.02.390
Pande V, Pandey SC, Sati D, Pande V, Samant M (2020) Bioremediation: an emerging effective approach towards environment restoration. Environ Sustain 3:91–103. https://doi.org/10.1007/s42398-020-00099-w
Bilal M, Rasheed T, Nabeel F, Iqbal HM, Zhao Y (2019) Hazardous contaminants in the environment and their laccase-assisted degradation—a review. J Environ Manag 234:253–264. https://doi.org/10.1016/j.jenvman.2019.01.001
Dasgupta J, Sikder J, Chakraborty S, Curcio S, Drioli E (2015) Remediation of textile effluents by membrane based treatment techniques: a state of the art review. J Environ Manag 147:55–72. https://doi.org/10.1016/j.jenvman.2014.08.008
Garcia-Garcia JD, Sanchez-Thomas R, Moreno-Sanchez R (2016) Bio-recovery of nonessential heavy metals by intra-and extracellular mechanisms in free-living microorganisms. Biotechnol Adv 34:859–873. https://doi.org/10.1016/j.biotechadv.2016.05.003
Bilal M, Rasheed T, Sosa-Hernández J, Raza A, Nabeel F, Iqbal H (2018) Biosorption: an interplay between marine algae and potentially toxic elements—a review. Mar Drugs 16:65. https://doi.org/10.3390/md16020065
Das S, Dash HR, Chakraborty J (2016) Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl Microbiol Biotechnol 100:2967–2984. https://doi.org/10.1007/s00253-016-7364-4
Lu TQ, Mao SY, Sun SL, Yang WL, Ge F, Dai YJ (2016) Regulation of hydroxylation and nitro reduction pathways during metabolism of the neonicotinoid insecticide imidacloprid by Pseudomonas putida. J Agric Food Chem 64:4866–4875. https://doi.org/10.1021/acs.jafc.6b01376
Mohammed YMM, Badawy MEI (2017) Biodegradation of imidacloprid in liquid media by an isolated wastewater fungus Aspergillus terreus YESM3. J Environ Sci Heal B 52:752–761. https://doi.org/10.1080/03601234.2017.1356666
Legradi JB, Di Paolo C, Kraak MH, Van der Geest HG, Schymanski EL, Williams AJ (2018) An ecotoxicological view on neurotoxicity assessment. Environ Sci Eur 30:46. https://doi.org/10.1186/s12302-018-0173-x
Wang Z, Wang Y, Gong F, Zhang J, Hong Q, Li S (2010) Biodegradation of carbendazim by a novel actinobacterium Rhodococcusjialingiae djl-6-2. Chemosphere 81:639–644. https://doi.org/10.1016/j.chemosphere.2010.08.040
Baybakova EV, Elena EN, Maria NB, Irina RG, Margarita DK, Galina AS (2020) Influence of fungicides on toxigenic properties of phytopathogenic fungi. BIO Web Conf 23:03006. https://doi.org/10.1051/bioconf/20202303006
Miller RNG, Alves GSC, Van Sluys MA (2017) Plant immunity: unravelling the complexity of plant responses to biotic stresses. Ann Bot 119:681–687. https://doi.org/10.1093/aob/mcw284
Efe D (2020) Potential plant growth promoting bacteria with heavy metal resistance. Curr Microbiol. https://doi.org/10.1007/s00284-020-02208-8
Shahid M, Khan MS (2017) Assessment of glyphosate and quizalofop mediated toxicity to greengram [Vignaradiata(L.) Wilczek], stress abatement and growth promotion by herbicide tolerant Bradyrhizobium and Pseudomonas species. Int J Curr Microbiol Appl Sci 6:3001–3016. https://doi.org/10.20546/ijcmas.2017.612.351
Lloyd JR, Lovley DR, Macaskie LE (2003) Biotechnological application of metal reducing microorganisms. Adv Appl Microbiol. https://doi.org/10.1016/S0065-2164(03)53003-9
Williams GP, Gnanadesigan M, Ravikumar S (2012) Biosorption and biokinetic studies of halobacterial strains against Ni2+, Al3+ and Hg2+metal ions. Bioresour Technol 107:526–529. https://doi.org/10.1016/j.biortech.2011.12.054
Roy T, Das N (2017) Isolation, characterization and identification of two methomyl degrading bacteria from a pesticide-treated crop field in West Bengal, India. Microbiology 86:753–764. https://doi.org/10.1134/S0026261717060145
Yadav S, Verma SK, Singh Chaudhary H (2015) Isolation and characterization of organophosphate pesticides degrading bacteria from contaminated agricultural soil. Online J Biol Sci 15:113–125
Dahiya P, Purkayastha S (2012) Phytochemical screening and antimicrobial activity of some medicinal plants against multi-drug resistant bacteria from clinical isolates. Indian J Pharm Sci 74:443–450. https://doi.org/10.4103/0250-474X.108420
Andrews JM (2001) Determination of minimum inhibitory concentrations. J Antimicrob Chemother 48:5–16. https://doi.org/10.1093/jac/48.suppl_1.5
de Silva A, de Carvalho AL, de Souza MAR, Dias SA, da Silva PMT, Filho RG et al (2012) Heavy metal tolerance (Cr, Ag and Hg) in bacteria isolated from sewage. Braz J Microbiol 43:1620–1631. https://doi.org/10.1590/S1517-838220120047000400
Liu SL, Yao K, Jia DY, Zhao N, Lai W, Yuan HY (2012) A pretreatment method for HPLC analysis of cypermethrin in microbial degradation systems. J Chromatogr Sci 50:469–476. https://doi.org/10.1093/chromsci/bms030
Sykora P, Čeplíková V, Foltýnová Z, Horniak L, Ebringer L (1991) Elimination of plasmids pKM 101 and F’lac from Salmonella typhimurium and Escherichia coli by bis ammonium salt. Folia Microbiol 36:240–245. https://doi.org/10.1007/BF02814355
Sivashankari TR, Sudha K, Barathi S, Karthikeyan V (2016) Efficacy of plasmid curing agent on Streptomyces longsporesflavns. Afr J Microbiol Res 10:616–625. https://doi.org/10.5897/AJMR2015.7675
Roy T, Bandopadhyay A, Sonawane P, Majumdar S, Mahapatra N, Alam S, Das N (2018) Bio-effective disease control and plant growth promotion in lentil by two pesticide degrading strains of Bacillus spp. Biol Control 127:55–63. https://doi.org/10.1016/j.biocontrol.2018.08.018
Bandopadhyay A, Roy T, Das N (2018) Isolation of some soil bacteria showing potentiality for disease control, growth enhancement and pesticide degradation in Vigna unguiculata L. Plant Arch 18:79–88
Pietrzak D, Kania J, Kmiecik E, Malina G, Wator K (2020) Fate of selected neonicotinoid insecticides in soil-water systems: current state of the art and knowledge gaps. Chemosphere 255:126981. https://doi.org/10.1016/j.chemosphere.2020.126981
Banerjee I, Tripathi SK, Sinha Roy A, Sengupta P (2014) Pesticide used pattern among farmers in rural district of West Bengal, India. J Nat Sci Biol Med 5:313–316. https://doi.org/10.4103/0976-9668.136173
Castillo JM, Casas J, Romero E (2011) Isolation of an endosulfan degrading bacterium from a coffee farm soil: persistence and inhibitory effect on its biological functions. Sci Total Environ 412:20–27. https://doi.org/10.1016/j.scitotenv.2011.09.062
Ahemad M, Khan SM (2012) Effects of pesticides on plant growth promoting traits of Mesorhizobium strain MRC4. J Saudi Soc Agric Sci 11:63–71. https://doi.org/10.1016/j.jssas.2011.10.001
Panda J, Kanjilal T, Das S (2018) Optimized biodegradation of carcinogenic fungicide carbendazim by Bacillus licheniformis JTC-3 from agro-effluent. Biotechnol Res Innov 2:45–57. https://doi.org/10.1016/j.biori.2017.10.004
Rani R, Kumar V (2017) Endosulfan degradation by selected strains of plant growth promoting rhizobacteria. Bull Environ Contam Toxicol 99:138–145. https://doi.org/10.1007/s00128-017-2102-x
Tang W (2017) Research progress of microbial degradation of organophosphorus pesticides. Prog Appl Microbiol 1:29–35
Ekram MA, Sarker I, Rahi MS, Rahman MA, Saha AK, Abu Reza M (2020) Efficacy of soil borne Enterobacter sp. for carbofuran degradation: HPLC quantitation of degradation rate. J Basic Microbiol 60:1–10. https://doi.org/10.1002/jobm.201900570
Bhatt P, Huang Y, Zhan H, Chen S (2019) Insight into microbial applications for the biodegradation of pyrethroid insecticides. Front Microbiol 10:1778. https://doi.org/10.3389/fmicb.2019.01778
Krishnasamy L, Sundaram CS, Sivakumar J (2019) Biodegradation of pesticides from the isolated microbial flora of crop field contaminated soil. Res J Life Bioinform Pharm Chem Sci 5:150–163. https://doi.org/10.26479/2019.0502.12
Roane TM, Pepper IL, Miller RM (2005) Microbial remediation of metals. In: Crawford RL, Crawford DL (eds) Bioremediation: principles and applications. Cambridge University Press, Cambridge, p 312
Rahman Z, Singh VP (2018) Assessment of heavy metal contamination and Hg-resistant bacteria in surface water from different regions of Delhi, India. Saudi J Biol Sci 25:1687–1695. https://doi.org/10.1016/j.sjbs.2016.09.018
Patel PR, Shaikh SS, Sayyed RZ (2016) Dynamism of PGPR in bioremediation and plant growth promotion in heavy metal contaminated soil. Indian J Exp Biol 54:286–290
Bandopadhyay A, Bhattacharya SK, Das N (2019) Biocontrol and growth promoting potential of eight PGPFs on jute and sunnhemp. J Soils Crops 29:243–250
Pietro-Souza W, de Campos PF, Mello IS, Stachack FFF, Terezo AJ, da Cunha CN et al (2020) Mercury resistance and bioremediation mediated by endophytic fungi. Chemosphere 240:124874. https://doi.org/10.1016/j.chemosphere.2019.124874
Malik A, Aleem A (2011) Incidence of metal and antibiotic resistance in Pseudomonas spp. from the river water, agricultural soil irrigated with wastewater and groundwater. Environ Monit Assess 178:293–308. https://doi.org/10.1007/s10661-010-1690-2
Pattnaik S, Dash D, Mohapatra S, Pattnaik M, Marandi AK, Das S, Samantaray DP (2020) Improvement of rice plant productivity by native Cr(VI) reducing and plant growth promoting soil bacteria Enterobacter cloacae. Chemosphere 240:124895. https://doi.org/10.1016/j.chemosphere.2019.124895
Capkin E, Terzi E, Altinok I (2015) Occurrence of antibiotic resistance genes in culturable bacteria isolated from Turkish trout farms and their local aquatic environment. Dis Aquat Organ 114:127–137. https://doi.org/10.3354/dao02852
Alaali Z, Thani ASB (2020) Patterns of antimicrobial resistance observed in the Middle East: Environmental and health care retrospectives. Sci Total Environ 740:140089. https://doi.org/10.1016/j.scitotenv.2020.140089
Anjum R, Grohmann E, Malik A (2011) Molecular characterization of conjugative plasmids in pesticide tolerant and multi-resistant bacterial isolates from contaminated alluvial soil. Chemosphere 84:175–181. https://doi.org/10.1016/j.chemosphere.2011.02.002
Trevors JT (1986) Plasmid curing in bacteria. FEMS Microbiol Rev 32:149–157. https://doi.org/10.1111/j.1574-6968.1986.tb01189.x
Dastidar SG, Poddar R, Kumar R, Chakrabarty AN (1977) Incidence and elimination of R plasmids in Vibrio cholerae. Antimicrob Agents Chemother 11:1079–1080. https://doi.org/10.1128/AAC.11.6.1079
Molina-Aja A, García-Gasca A, Abreu-Grobois A, Bolán-Mejía C, Roque A, Gomez-Gil B (2002) Plasmid profiling and antibiotic resistance of Vibrio strains isolated from cultured Penaeid shrimp. FEMS Microbiol Lett 213:7–12. https://doi.org/10.1111/j.1574-6968.2002.tb11278.x
Nies DH, Brown NL (1998) Two component system in regulation of heavy metal resistance. In: Silver S, Walden W (eds) Metal ions in gene regulation. Springer, New York, pp 77–103. https://doi.org/10.1007/978-1-4615-5993-1_4
Agarwal M, Rathore RS, Jagoe C, Chauhan A (2019) Multiple lines of evidences reveal mechanisms underpinning mercury resistance and volatilization by Stenotrophomonas sp. MA5 isolated from the Savannah river site (SRS), USA. Cells 8:309. https://doi.org/10.3390/cells8040309
Malik A, Çelik EK, Bohn C, Böckelmann U, KnobelK GE (2008) Detection of conjugative plasmids and antibiotic resistance genes in anthropogenic soils from Germany and India. FEMS Microbiol Lett 279:207–216. https://doi.org/10.1111/j.15746968.2007.01030.x
Maheswari UA, Nuni A, Shreevidya R (2010) Evaluation of antibacterial activity of Boerhaavia diffusa L. leaves. Int J Green Pharm 4:75–78. https://doi.org/10.22377/ijgp.v4i2.123
Hawkins NJ, Bass C, Dixon A, Neve P (2019) The evolutionary origins of pesticide resistance. Biol Rev 94:135–155. https://doi.org/10.1111/brv.12440
Acknowledgements
The work was financially supported by Department of Science and Technology (West Bengal), India [Memo no. 757 (Sanc)/ST/P/S&T/1G-15/2014]. We acknowledge the help of Mr. S. Biswas, Central Instrumental Facility, Bose Institute, Kankurgachhi, Kolkata, India for HPLC analysis.
Author information
Authors and Affiliations
Contributions
ND and SM developed the concept. ND and SM designed the experiment. TR performed the experiments. ND, AB, and SM analyzed the data. ND, TR, and CP interpreted the results. TR, AB, CP, and ND wrote the manuscript. ND overall supervised the work.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that there are no conflicts of interest in the present investigation.
Ethics Approval
Not applicable.
Consent to Participate
Not Applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roy, T., Bandopadhyay, A., Paul, C. et al. Role of Plasmid in Pesticide Degradation and Metal Tolerance in Two Plant Growth-Promoting Rhizobacteria Bacillus cereus (NCIM 5557) and Bacillus safensis (NCIM 5558). Curr Microbiol 79, 106 (2022). https://doi.org/10.1007/s00284-022-02793-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00284-022-02793-w