Abstract
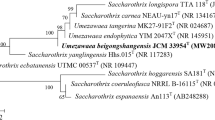
A novel actinobacterium, YIM 132087T, isolated from Lepraria sp. lichen collected from Yunnan province, south-west PR China. Cells are Gram-stain-positive, catalase-positive and oxidase-negative, aerobic, non-motile and short rod-shaped. Colonies are asporogenous, circular and white brown in colour. Optimal growth occured at 15−35 °C (optimum 28 °C), at pH 5.0−9.0 (optimum pH 6.0), and in the presence of 3% NaCl (w/v). The DNA G+C content of strain YIM 132087T based on the draft genome sequence was 71.3 mol%. Phylogenetic analysis based on 16S rRNA gene sequences suggested that strain YIM 132087T belonged to the genus Nakamurella and exhibited high levels of 16S rRNA gene sequence similarity with Nakamurella endophytica CGMCC 4.7038T (97.9%) and Nakamurella intestinalis NBRC 111844T (97.2%). The DNA–DNA hybridization values between strain YIM 132087T and its closest relatives are lower than 26%. Strain YIM 132087T had meso-diaminopimelic acid as the diagnostic cell-wall diamino acid, and MK-8(H4) as the predominant menaquinone. Predominant cellular fatty acids (> 10%) were iso-C16:0, iso-C15:0, C16:0 and anteiso-C15:0. The polar lipid profile were found to be diphosphatidylglycerol, phosphatidylmethylethanolamine, phosphatidylethanolamine, phosphatidylinositol, three unknown phospholipids, one unknown aminophospholipid and one unknown lipid. Based on phenotypic, phylogenetic and chemotaxonomic analysis, strain YIM 132087T belongs to the genus Nakamurella and represents a novel species of the genus Nakamurella, for which the name Nakamurella albus sp. nov., with type strain YIM 132087T (=CGMCC 4.7629T =NBRC 114017T), is proposed.

Similar content being viewed by others
References
Yoshimi Y, Hiraishi A, Nakamura K (1996) Isolation and characterization of Microsphaera multipartita gen. nov., sp. nov., a polysaccharide accumulating gram-positive bacterium from activated sludge. Int J Syst Bacteriol 46:519–525
Tao TS, Yue YY, Chen WX, Chen WF et al (2004) Proposal of Nakamurella gen. nov. as a substitute for the bacterial genus Microsphaera Yoshimi et al. 1996 and Nakamurellaceae fam. nov. as a substitute for the illegitimate bacterial family Microsphaeraceae Rainey et al. 1997. Int J Syst Evol Microbiol 54:999–1000
Yoon JH, Kang SJ, Jung SY, Oh TK (2007) Humicoccus flavidus gen. nov., sp. nov., isolated from soil. Int J Syst Evol Microbiol 57:56–59
Kim KK, Lee KC, Lee JS (2012) Nakamurella panacisegetis sp. nov. and proposal for reclassification of Humicoccus flavidus Yoon et al., 2007 and Saxeibacter lacteus Lee et al., 2008 as Nakamurella flavida comb. nov. and Nakamurella lactea comb. nov. Syst Appl Microbiol 35:291–296
Lee SD, Park SK, Yun YW, Lee DW (2008) Saxeibacter lacteus gen. nov., sp. nov., an actinobacterium isolated from rock. Int J Syst Evol Microbiol 58:906–909
Tuo L, Li FN, Pan Z, Lou I, Guo M et al (2016) Nakamurella endophytica sp. nov., a novel endophytic actinobacterium isolated from the bark of Kandelia candel. Int J Syst Evol Microbiol 66:1577–1582
França L, Albuquerque L, Zhang DC, Nouioui I, Klenk HP et al (2016) Nakamurella silvestris sp. nov., an actinobacterium isolated from alpine forest soil. Int J Syst Evol Microbiol 66:5460–5464
Kim SJ, Cho H, Joa JH, Hamada M, Ahn JH et al (2017) Nakamurella intestinalis sp. nov., isolated from the faeces of Pseudorhynchus japonicus. Int J Syst Evol Microbiol 67:2970–2974
Hayakawa M, Nonomura H (1987) Humic acid-vitamin agar, a new medium for the selective isolation of soil acinomycetes. J Ferment Technol 65:501–509
Liu CB, Jiang Y, Wang XY, Chen DB, Chen X, Wang LS, Han L, Huang XS, Jiang CL (2017) Diversity, antimicrobial activity, and biosynthetic potential of cultivable actinomycetes with lichen symbiosis. Microb Ecol 74:570–584
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBiocloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstucting phylogenetic trees. Mol Biol Evol 4:406–425
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrat. Evolution 39:783–791
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Leifson E (1960) Atlas of bacterial flagellation. Academic Press, New York
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703–703
Dong XZ, Cai MY (2001) Determination of biochemical properties. In: Dong XZ, Cai MY (eds) Manual for the systematic identification of general bacteria. Science Press, Beijing (in Chinese), pp 370–398
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov. sp. nov., a novel member of the family 'oxalobacteraceae' isolated from china. Int J Syst Evol Microbiol 55:1149–1153
Denner EBM, Paukner S, Kämpfer P, Moore ERB, Abraham WR, Busse HJ, Wanner G, Lubitz W (2001) Sphingomonas pituitosa sp. nov., an exopolysaccharide-producing bacterium that secretes an unusual type of sphingan. Int J Syst Evol Microbiol 51:827–841
Kuykendall LD, Roy MA, O'Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Newark, DE: MIDI
Tindall BJ (1990) Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol Lett 66:199–202
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Tang SK, Wang Y, Chen Y, Lou K, Cao LL et al (2009) Zhihengliuella alba sp. nov., and emended description of the genus Zhihengliuella. Int J Syst Evol Microbiol 59:2025–2032
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Christensen H, Angen O, Mutters R, Olsen JE, Bisgaard M (2000) DNA–DNA hybridization determined in microwells using covalent attachment of DNA. Int J Syst Evol Microbiol 50:1095–1102
Li R, Li Y, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Li D, Liu CM, Luo R, Sadakane K, Lam TW (2015) MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676
Acknowledgements
This research was funded by National Natural Science Foundation of China (31460005) and Major research project of Guangxi for science and technology (AA18242026).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of strain YIM 132087T is MN317339 and the genome sequence is WLYK00000000.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, LQ., An, DF., Zhang, K. et al. Nakamurella albus sp. nov.: A Novel Actinobacterium Isolated from a Lichen Sample. Curr Microbiol 77, 1896–1901 (2020). https://doi.org/10.1007/s00284-020-01928-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-020-01928-1