Abstract
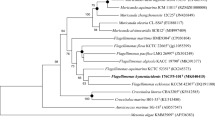
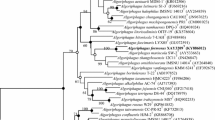
A Gram-stain-negative, strictly aerobic and moderately halophilic bacterium, designated strain AsT0115T, was isolated from a red marine alga, Asparagopsis taxiformis. Cells were non-motile rods showing oxidase-positive and catalase-negative activities. Growth was observed at 15–37 C (optimum, 25 C) and pH 6.5–7.5 (optimum, pH 7.0) and in the presence of 0–11.0% (w/v) NaCl (optimum, 2.0–3.0%). Strain AsT0115T contained iso-C15:0 and iso-C15:1 G as the predominant fatty acids. Menaquinone-6 was identified as the sole isoprenoid quinone. Major polar lipids consisted of phosphatidylethanolamine, an identified aminolipid and three unidentified polar lipids. The G+C content of the genomic DNA calculated from the whole genome sequence was 41.7 mol%. Strain AsT0115T was most closely related to Flagellimonas pacifica sw169T and Flagellimonas flava DSM 22638T with 96.5% and 95.6% 16S rRNA gene sequence similarities, respectively. Phylogenetic analysis based on 16S rRNA gene sequences showed that strain AsT0115T formed a phyletic lineage with F. pacifica sw169T within the genus Flagellimonas. Phenotypic, chemotaxonomic, and phylogenetic features clearly suggested that strain AsT0115T represents a novel species of the genus Flagellimonas, for which the name Flagellimonas algicola sp. nov. is proposed. The type strain is AsT0115T (= KACC 19790T = JCM 32942T).

Similar content being viewed by others
References
Bae SS, Kwon KK, Yang SH, Lee HS, Kim SJ, Lee JH (2007) Flagellimonas eckloniae gen. nov., sp. nov., a mesophilic marine bacterium of the family Flavobacteriaceae, isolated from the rhizosphere of Ecklonia kurome. Int J Syst Evol Microbiol 57:1050–1054
Yoon BJ, Oh DC (2012) Spongiibacterium flavum gen. nov., sp. nov., a member of the family Flavobacteriaceae isolated from the marine sponge Halichondria oshoro, and emended descriptions of the genera Croceitalea and Flagellimonas. Int J Syst Evol Microbiol 62:1158–1164
Gao X, Zhang Z, Dai Z, Zhang XH (2015) Spongiibacterium pacificum sp. nov., isolated from seawater of South Pacific Gyre and emended description of the genus Spongiibacterium. Int J Syst Evol Microbiol 65:154–158
Choi S, Lee JH, Kang JW, Choe HN, Seong CN (2018) Flagellimonas aquimarina sp. nov., and transfer of Spongiibacterium flavum Yoon and Oh 2012 and S. pacificum Gao, 2015 to the genus Flagellimonas Bae et al. 2007 as Flagellimonas flava comb. nov. and F. pacifica comb. nov., respectively. Int J Syst Evol Microbiol 68:3266–3272
Jeong SE, Kim KH, Lhee D, Yoon HS, Quan ZX, Lee EY, Jeon CO (2019) Oceaniradius stylonematis gen. nov., sp. nov., isolated from a red alga, Stylonema cornu-cervi. Int J Syst Evol Microbiol 69:1967–1973
Jung HS, Jeong SE, Chun BH, Quan ZX, Jeon CO (2019) Rhodophyticola porphyridii gen. nov., sp. nov., isolated from a red alga, Porphyridium marinum. Int J Syst Evol Microbiol 69:1656–1661
Feng T, Kim KH, Jeong SE, Kim W, Jeon CO (2018) Aquicoccus porphyridii gen. nov., sp. nov., isolated from a small marine red alga, Porphyridium marinum. Int J Syst Evol Microbiol 68:283–288
Murphy CD, Moore RM, White RL (2000) Peroxidases from marine microalgae. J Appl Phycol 12:507–513
Kawasaki K, Kamagata Y (2017) Phosphate-catalyzed hydrogen peroxide formation from agar, gellan, and κ-carrageenan and recovery of microorganisms cultivability by catalase and pyruvate. Appl Environ Microbiol 83:e01366–e1417
Lee Y, Jeon CO (2018) Solitalea longa sp. nov., isolated from freshwater and emended description of the genus Solitalea. Int J Syst Evol Microbiol 68:2826–2831
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Gomori G (1955) Preparation of buffers for use in enzyme studies. Methods Enzymol 1:138–146
Lányi B (1987) Classical and rapid identification methods for medically important bacteria. Methods Microbiol 19:1–67
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P (ed) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, Tang J (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Minnikin DE, O'Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark
Minnikin DE, Patel PV, Alshamaony L, Goodfellow M (1977) Polar lipid composition in the classification of Nocardia and related bacteria. Int J Syst Bacteriol 27:104–117
Stackebrandt E, Ebers J (2006) Taxonomic parameter revisited: tarnished gold standards. Microbiol Today 33:152–155
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Rosselló-Móra R, Amann R (2015) Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38:209–216
Acknowledgements
This work was supported by the Program for Collection of Domestic Biological Resources of the National Institute of Biological Resources (NIBR201902203) of the Ministry of Environment (MOE) and Future Planning and the Strategic Initiative for Microbiomes in the Ministry of Agriculture, Food, and Rural Affairs (as part of the multi-ministerial) Genome Technology to Business Translation Program, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing financial conflicts of interests.
Ethical Approval
The authors have declared that no ethical issues exist.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank accession numbers for the 16S rRNA gene and genome sequences of strain S-16T are MK391377 and VCNI00000000, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, J., Kim, K.H., Chun, B.H. et al. Flagellimonas algicola sp. nov., Isolated from a Marine Red Alga, Asparagopsis taxiformis. Curr Microbiol 77, 294–299 (2020). https://doi.org/10.1007/s00284-019-01821-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-019-01821-6